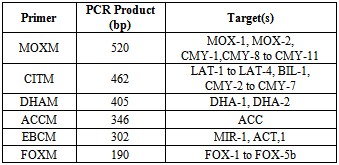
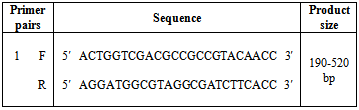
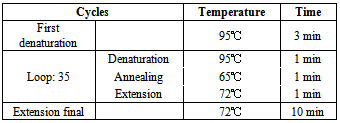
| [1] | Shacheraghi F, Shakibaie MR, and Noveiri H. Molecular Identification of ESBL Genes blaGES-1, blaVEB-1, blaCTX-M blaOXA-1, blaOXA-4, blaOXA-10 and blaPER-1 in Pseudomonas aeruginosa Strains Isolated from Burn Patients by PCR, RFLP and Sequencing Techniques. World Academy of Science, Engineering and Technology. 2010; 37: 1189-1193. |
| [2] | Juan C, Macia MD, Gutierrez O, Vidal C, Perez JL, and Oliver A. Molecular Mechanisms of ß-Lactam Resistance Mediated by AmpC Hyperproduction in Pseudomonas aeruginosa Clinical Strains. Antimicrobial agents and chemotherapy. 2005; 49(11): 4733–4738. |
| [3] | Zavascki A.P, Carvalhaes CG, Picao RC, and Gales A.C. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacterbaumannii: resistance mechanisms and implications for therapy. Expert Review of Anti-Infective Therapy. 2012; 8(1): 71-93. |
| [4] | Maatallah M, Cheriaa J, Backhrouf A, Iversen A, Grundmann H, et al. Population Structure of Pseudomonas aeruginosa from Five Mediterranean Countries: Evidence for Frequent Recombination and Epidemic Occurrence of CC235. 2011; 6(10): e25617. |
| [5] | McGowan JE. Resistance in nonfermenting Gram-negative bacteria: multidrug resistance to the maximum. The American Journal of Medicine. 2006; 119(6): S29–S36. |
| [6] | Strateva T, Ouzounova-Raykova V, Markova B, Todorova A, Marteva-Proevska Y, and Mitov I. Problematic clinical isolates of Pseudomonas aeruginosa from the university hospitals in Sofia, Bulgaria: current status of antimicrobial resistance and prevailing resistance mechanisms. Journal of Medical Microbiology. 2007; 56: 956–963. |
| [7] | Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection. 2011; 18(3): 268–281. |
| [8] | Bonomo RA, Scabo D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clinical Infectious Diseases. 2006; 43(2): 49-56. |
| [9] | Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: Our Worst Nightmare? Clinical Infectious Diseases. 2002; 34(5): 634–640. |
| [10] | Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clinical Microbiology and Infection. 2007; 13: 5–18. |
| [11] | Philippon A, Arlet G, and Jacoby GA. Plasmid-Determined AmpC-Type ß-Lactamases .Antimicrobial agents and chemotherapy. 2002; 46(1): 1–11. |
| [12] | Lodge JM, Minchin SD, Piddock LJV, and Busby SJW. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC ß-lactamase. Biochem. J. 1990; 272: 627-631. |
| [13] | Bennett PM. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. British Journal of Pharmacology. 2008; 153(S1): S347–S357. |
| [14] | Chroma M, Kolar M. Genetic methods for detection of antibiotic resistance: focus on extended-spectrum β-lactamases. 2010; 154(4): 289–296. |
| [15] | Livermore DM, Woodford N. The ß-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbial. 2006; 14(9):413-420. |
| [16] | Ambler RP. The structure of ß-lactamases. Philosophical transaction, the Royal society, Lond B. biological sciences. 1980; 289(1036): 321-331. |
| [17] | Bush K, Jacoby GA, and Medeiros AA. A functional classification scheme for ß-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995; 39(6): 1211–1233. |
| [18] | Sanders CC. Chromosomal cephalosporinases responsible for multiple resistance to newer ß-lactam antibiotics. Annual Review of Microbiology. 1987; 41: 573–593. |
| [19] | Giwercman B, Lambert PA, Rosdahl VT, Shand GT, and Hoiby N. Rapid emergence of resistance in Pseudomonas aeruginosa in cystic fibrosis patients due to in-vivo selection of stable partially derepressed ß-lactamase producing strains. Antimicrobial agents and chemotherapy. 1990; 26: 247–259. |
| [20] | Livermore DM. Clinical significance of beta-lactamase induction and stable derepression in gram-negative rods. Eur. J. Clin. Microbiol. 1987; 6: 439–445. |
| [21] | Livermore DM. ß-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 1995; 8: 557–584. |
| [22] | Jacoby GA .AmpC ß –lactamases. Clinical Microbiology. Rev. 2009; 22(1): 161-182. |
| [23] | Dumas JL, Delden CV, Perron K, and Kohler T. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. Federation of European Microbiological Societies Microbiol Lett. 2006; 254: 217–225. |
| [24] | Langaee TY, Gagnon L, and HuletskyA. Inactivation of the ampD Gene in Pseudomonas aeruginosa Leads to Moderate- Basal-Level and Hyperinducible AmpC ß-Lactamase Expression. Antimicrobial Agents and Chemotherapy. 2000; 44(3): 583–589. |
| [25] | Hancock REW, Speert DP. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resistance Updates. 2000; 3: 247–255. |
| [26] | Jacobs C, Frere JM, and Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible ß-lactam resistance in gram-negative bacteria. Cell Press. 1997; 88(6): 823–832. |
| [27] | Wiedemann B, Dietz H, and Pfeifle D. Induction of ß-lactamase in Enterobacter cloacae. Clinical Infectious Diseases. 1988; 27(1): S42–S47. |
| [28] | Langaee TY, Dargis M, and HuletskyA. An ampD gene in Pseudomonas aeruginosa encodes a negative regulator of AmpC ß-lactamase expression. Antimicrob. Agents Chemother. 1998; 42:3296–3300. |
| [29] | Lodge J, Busby S, and Piddock L. Investigation of the Pseudomonas aeruginosaampR gene and its role at the chromosomal ampC promoter. Federation of European Microbiological Societies Microbiol Lett. 1993; 111: 315–320. |
| [30] | Manoharan A, Sugumar M, Kumar A, Jose H, and Mathai D. Phenotypic & molecular characterization of AmpC β-lactamases among Escherichia coli, Klebsiella spp. & Enterobacter spp. from five Indian Medical Centers. Indian J Med Res. 2012; 135: 359-364. |
| [31] | Mansouri S, Chitsaz M, Haji-hosseini R, Mirzaee M, and Ghini MH. Determining resistance of E.coli clinical isolates producing AmpC betalactamase based on their phenotypic and genotypic properties. Shahed university J. 1388; 19(80): 1-11. |
| [32] | Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection. 2011; 18(3): 268-281. |
| [33] | Jiang X, Zhang Z, Li M, Zhou D, Ruan F, and Lu Y. Detection of Extended-Spectrum ß-Lactamases in Clinical Isolates of Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2006; 50(9): 2990–2995. |
| [34] | Tassios PT, Gennimata V, L. Spaliara-Kalogeropoulou, D. Kairis, C. Koutsia, A. C. Vatopoulos, and N. J. Legakis. 1997. Multiresistant Pseudomonas aeruginosaserogroup O: 11 outbreak in an intensive care unit. Clinical Microbiology and Infection. 3(6): 621–628. |

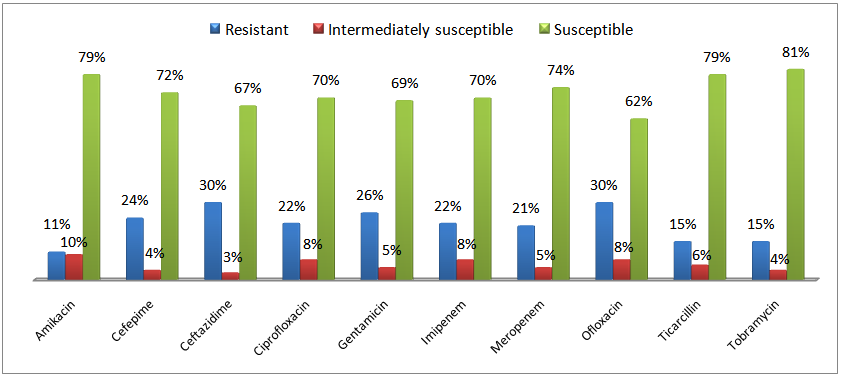
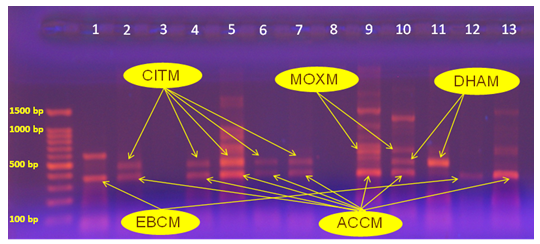
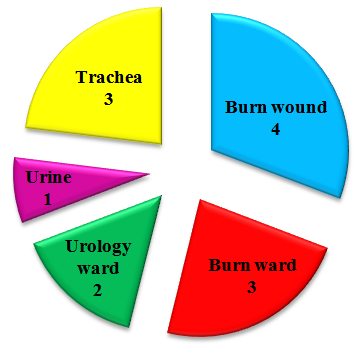
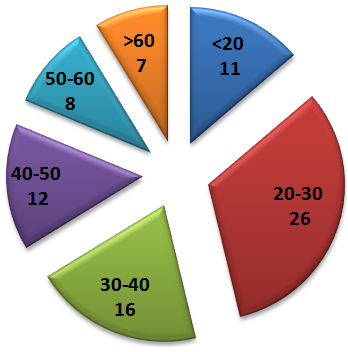
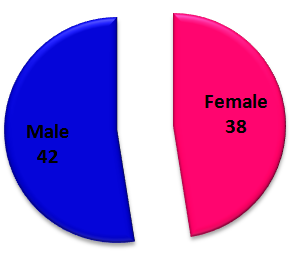
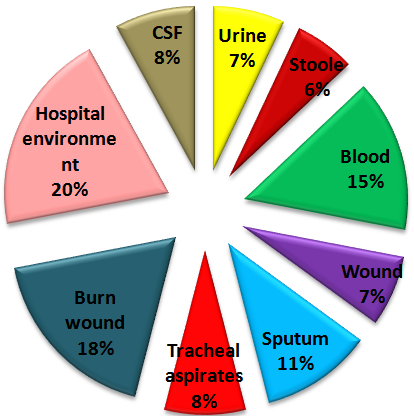
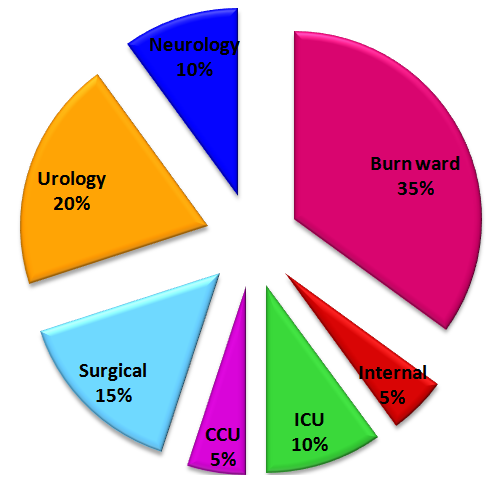
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML