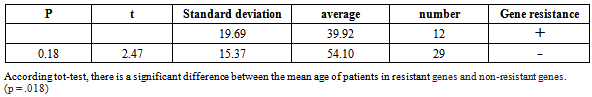Milad Farokhzad1, Bahareh Rahimian Zarif2
1Department of biology, Kurdistan science and research branch, Islamic Azad University, Sanandaj, Iran
2Department of biology, Sanandaj branch, Islamic Azad University, Sanandaj, Iran
Correspondence to: Bahareh Rahimian Zarif, Department of biology, Sanandaj branch, Islamic Azad University, Sanandaj, Iran.
| Email: |  |
Copyright © 2014 Scientific & Academic Publishing. All Rights Reserved.
Abstract
Literature review and objectives: Klebsiella pneumoniae is one of the most important causes of nosocomial infections and antibiotic resistance. Integron is one of the genetic movable elements that can carry different antibiotic resistance genes. Here, the class 1 integron role in the creation and transfer of antibiotic resistance is very important. The purpose of this study is to isolate K. pneumoniae from hospitalized patients of Tohid Hospital in Sanandaj and molecular analysis of class 1 and 2 integrase genes and gene cassettes CS1 and CS2 and also determining susceptibility or resistance to different antibiotics. Materials and methods: 120Klebsiella samples were collected from Tohid hospital in Sanandaj. Klebsiellas were tested by using standard methods and differential tests and 41 samples, 41 isolates of Klebsiella pneumoniae were detected from all the samples. Susceptibility testing was performed by Standard disk diffusion method according to CLSI method. Inorder to investigate the class 1 & 2 integrons frequency and also, gene cassettes CS1 & CS2, PCR method was applied. Findings: Of all the 120 klebsiella samples, 41wereKlebsiella pneumoniae. The result of antibiotic-sensetivity test showed that 100% of the strains were sensetive to imipenem and meropenem, 83.33% to gentamicin, 100% to ceftriaxone, 75% to amikacin, 100% to cefepime, 83.33% to ciprofloxacin, 100% to ceftazidime, 100% to cefotaxime and 100 % were resistant to Ztryvnam. Of the 41 resistant samples, in 12 strains (29.26%) integrase 1 and 5 strains (12.19%) gene cassette CS2 was observed, but in no strain, class 2 integrase and gene cassette CS2 were observed. Results: The results of the research indicated that class1 integrase gene frequency is high in strains which can play an important role in creation and transfer of antibiotic resistance; and this could be due to the indiscriminate use of antibiotics.
Keywords:
Klebsiella pneumoniae, Class 1 & 2 integrons, Multi-drug resistant, PCR
Cite this paper: Milad Farokhzad, Bahareh Rahimian Zarif, Studying Class 1 & 2 Integrons and Gene Cassettes CS1 & CS2 in Multi-drug Resistant Klebsiella Pneumonia Isolates of Clinical Samples Taken from Hospitalized Patients in Tohid Hospital of Sanandaj, in 2013, Advances in Life Sciences, Vol. 4 No. 2, 2014, pp. 59-64. doi: 10.5923/j.als.20140402.04.
1. Introduction
Klebsiella pneumoniae is an opportunistic pathogen causing urinary tract infection and pneumonia in people with weakened immune systems [1]. Klebsiella pneumoniae with 15 to18 percent, has the second ranking after Escherichia coli in creation of septicemia and urinary tract infections as well as hospital Gram-negative infections [1]. Urinary tract infections are the most common infections in all age groups whose lack of diagnosis and treatment results in hypertension, renal disorders, and early pregnancy and even abortion, and urinary tract disorders [2 & 3]. In the past few years throughout the world, resistance to several drugs in bacteria compared to more than two different class of antibiotics among hospital isolates has been increasing; This is termed Multidrug resistant or MDR, it is said that this phenomenon will unfortunately be worrying [4 & 5]. Studies show that regardless of the pattern of use of antibiotics, antibiotic resistance genes can be transferred between bacterial populations [6]. DNA elements such as transposons and Plasmids which are the location of antibiotic resistance genes have been the cause of transfer of the genes between different species of bacteria; these elements are called the integron.Integrons have been introduced asa new mechanism for publishing resistance genes among bacteria [7]. Horizontal transfer of integrons is the most successful way in the release of resistance genes and emergence of multi-resistant species. Antibiotic-resistance coding integrons have been divided into four classes; Most of the known integrons belong to class 1 and they contain Sullgene, Class 2 is in teh transposons of the Tn-7 and class 3 contains Metallo-beta-lactamase [8 & 9 & 10].Generally based on differences in the integrase gene, integrons are divided into different classes. The four distinct classes of the integrons responsible for multiple resistance are a genetic code of aspecial integrase each. Integrase genes include Int I1, Int I2, Int I3, and Int I4. Most of the isolated samples from patients and hospital environments with MDR conditions belong to class 1 which are located on plasmids or transposons [11].
2. Materials and Methods
Bacterial isolates and data collectionInsummer 1392 collected samples that had been frozen were cultured on blood agar and eosin methylene blue prepared by the Merck Company in Germany. Profile of each isolate which include dage, sex, history of previous illness, types of samples were recorded in atable. It should be noted that the isolated Klebsiella pneumoniae samples were kept in Muller Hinton Broth environment (Merck, Germany) with 18% glycerolat- 80° Cuntil usage.Disk diffusion testing: Ten antibiotic disks produced by (MAST Company-UK), as well as a number of other antibiotics produced by the company (Exir -Iran) were tested for all the samples.
2.1. Antibioticsareas Follows
Imipenem (10 micrograms), gentamicin (10 mg), ceftriaxone (30 micrograms), amikacin (30 mg), cefepime (30 micrograms), ciprofloxacin (5 micrograms), ceftazidime (30 micrograms), cefotaxime (30 micrograms), meropenem (10 micrograms) and aztreonam (30 mcg). Klebsiella pneumoniae bacterium was cultured in Broth vitro (Barin Heart Infusion BHI) and incubated at 37°C for 24 hours. After bacterial growth and creation of turbidity according to semi McFarl and standard, they were cultured using steel swab on Mueller Hinton Agar environment. After the disks, the plates were placed in temperature of 37°C for 24 hours. Susceptibilityor resistance to antibiotics were reported based on NCCLS. Then the samples that were resistant to at least three classes of antibiotics were considered as examples of multiple resistance (multidrug resistance).
2.2. DNA Extraction, Performing PCR
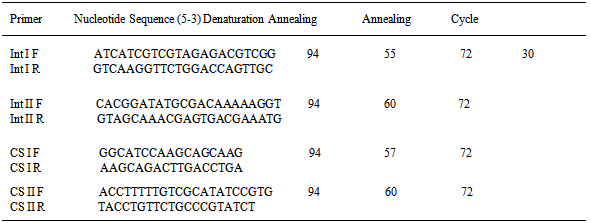
At this level phenol-chloroform method was used to extract DNA. After DNA extraction, each sample was maintained until PCR performance at a temperature of-20°C. To perform PCR primers sequences (Table 1) for a plurality of sequences of genesint I & int II, and also, the gene cassettes CS1 and CS2 were designed. PCR reactions were performedina volume of 50 microliter which contained 10 microliters of extracted DNA (1 microgram), 5 pmol of each primer per liter, 1.5 mmol per Liter MaCl2, 0.2mM per liter dNTPs and 1.5 unit of Taqenzyme. Proliferation of the intended pieces in the thermo cycler device is as follows:Table 1. Sequences and temperatures of primers used in the PCR
 |
| |
|
Electrophoresis of PCR products was done in Agarose gel of 1.5% and the voltage of 80 V foran hour. The gel was examined under UV light. The achieved bands compared to DNA size Marker of a thousand base pairs bp (Fermentas) were considered as the intended fragment of the genes.
3. Findings
Of all the 120 klebsiella samples, 41 were Klebsiella pneumoniae. The result of antibiotic-sensitivity test showed that 100% of the strains were sensitive to imipenem and meropenem, 83.33 % to gentamicin, 100% to ceftriaxone, 75% to amikacin, 100% to cefepime, 83.33 % to ciprofloxacin, 100% to ceftazidime, 100 % to cefotaxime and 100% were resistant to aztreonam. Of the 41 resistant samples, in 12 strains (29.26%) integrase 1 and 5 strains (12.19%) gene cassette CS2 was observed, but in any strain, class 2 integrase and gene cassette CS2 were not observed. In this study, 41 isolates of Klebsiella pneumoniae from Tohid Hospital in Sanandaj, were taken out. After performing PCR withspecific primers, 29.26 % of samples (12 samples) possessed geneint Iand 12. 19 % of the samples (5 samples) possessed gene cassette CS1; and Geneint II and gene cassette CS2 were not found.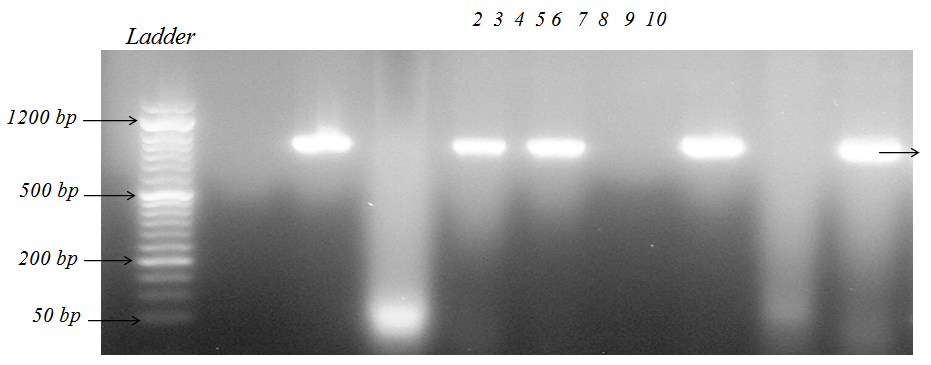 | Figure 1. PCR results of Class I integrongene of K. pneumoniae strains isolated from patients hospitalized in Tohid hospital in Sanandaj. (The firstplate marker bp 50 to 1500 bp, plates 3, 5, 6, 8 & 10 are positive class I integron gene and plates 2, 4, 7, 9 have reported negative results for class I integron gene) |
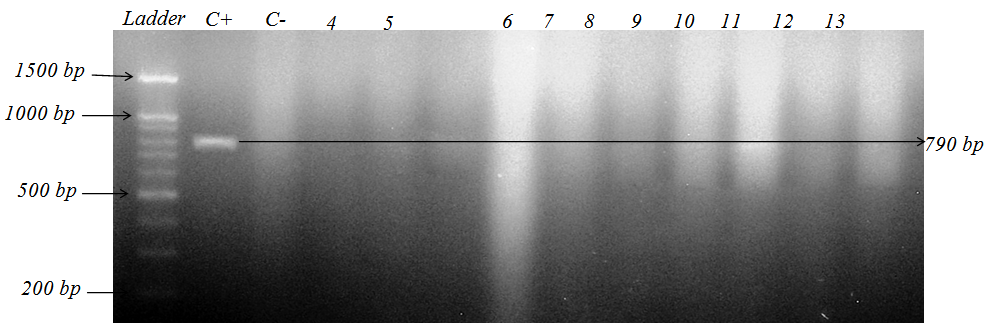 | Figure 2. PCR results of Class II integrongene of K. pneumoniae strains isolated from patients hospitalized in Tohid hospital in Sanandaj, (The first plate marker 200 bp to 1500 bp, the second plate Acinetobacter baumannii strains positive control and in plate three, negative control and class II integron gene cannot be seen therefore there is no CSII gene cassette |
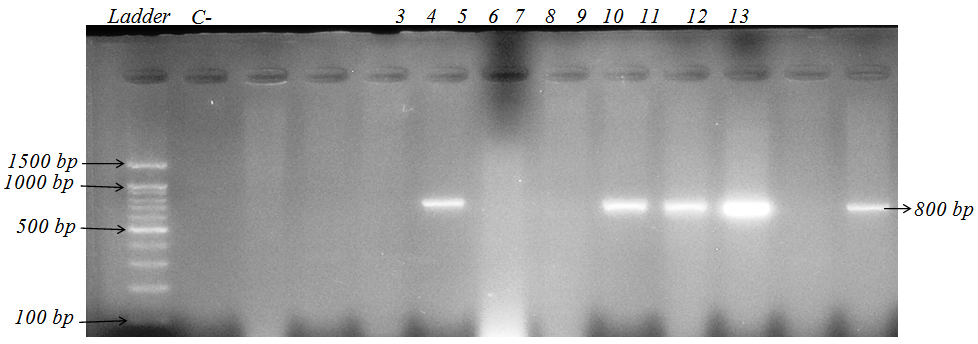 | Figure 3. PCR results CS1 gene cassette of K. pneumoniae strainsisolated from patients hospitalized in Tohid hospital in Sanandaj. (Plates 6, 9,10,11,13 contain gene cassette CS I. CS I bandsarevariable and this indicates the patients in the hospital have been contaminated with a specific clone of bacteria) |
For 41 isolates of Klebsiella pneumoniae, antibiotic sensitivity testing was performed, and the antibiotic resistance percentage of the isolated samples are as shown below in order: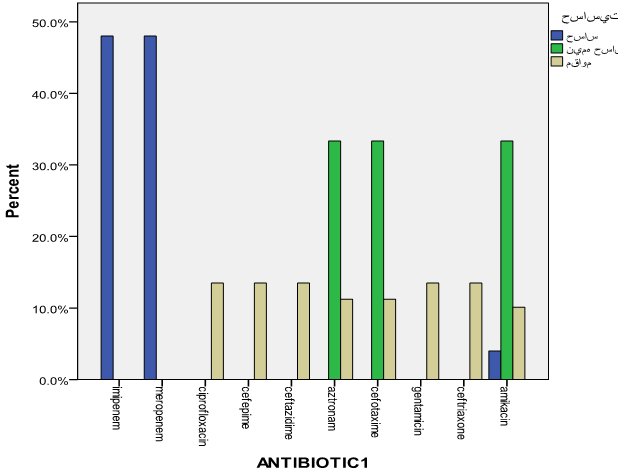 | Diagram 1. The frequency of antibiotic sensitivity |
4. Results and Discussions
Urinary tract infections in humans is considered one of the most important infections whose abundance is in second place after respiratory infection. However, Shah Cheraghi et.al. Reported the outbreak of Klebsiella pneumoniae isolates from urine samples greater than other urinary pathogens factors. In the present study 41 isolates of Klebsiella pneumoniae were taken out of a total of 175 urine samples from patients hospitalized in Tohid Hospital in Sanandaj, of these, 58. 54 % of isolates belonged to female patients and 41.46 % of the isolates were males. Khalili, et.al and also Ahangarzade Rezaee et.al have reported the results of positive urine culture in females 82.08 % and 55.55% and the results related to males 27.72% and 44.44% respectively. This indicates that women more than men are likely to developurinary tract infections particularly Klebsiella pneumoniae. Frequency of class I integrongenesin K. pneumoniae strains was 29.26 % which is consistent with the results reported by similar studies 34.02 % and 32.09 %. In a study conducted in a hospital in the United States in 2006, the outbreak of class I integron gene was reported 70 %; and in another study carried out in three hospitals in Australia in 2005 on Klebsiella producers of broad-spectrum beta-lactamase, the incidence of this gene was reported 73% which was not consistent with those of ours. This difference could be due to geographical regions and bacterial strains; thus the frequency percentage of integrons in different bacteria can vary, for example, in a report released in 2006 in China, the outbreak of Class I integronin Pseudomonas bacteria was reported 38% and in Amazon region it was 41.15%. In 2006, in 13 states of the United States of America the outbreak of integron gene among Escherichia coli and Klebsiella bacteria were studied using PCR; of the 320 bacteria isolates, 57% possessed class I inegron, of which 49% Escherichia coli bacteria and 70% Klebsiella pneumoniae bacteria possessed this gene. In this research, all the Klebsiella pneumoniae isolates possessing class I integron were resistant to cefepime, ceftazidime, aztreonam, cefotaxime, and ceftriaxone antibiotics. The reason for this resistance can be the presence of antibiotic resistance gene cassettes in class I integron gene. The antibiotic resistance level among isolates obtained from society and hospitals is increasing and this is a universal problem. Resistance in strains causing hospital infections is increasing which has led to many problems in curing the diseases. Regardless of the antibiotic consumption patterns, their resistant genes can be transferred among the bacterial population, meanwhile, the integron using the new mechanism of spread, transfer the resistance genes among bacteria. Horizontal transfer of integrons is the most successful way to release resistance genes and lead tothe emergence of species with multi-drug resistance (MDR).Table 2. Antibiotic susceptibility test of isolates possessing positive integron taken from Tohid Hospitalin Sanandaj in 2013
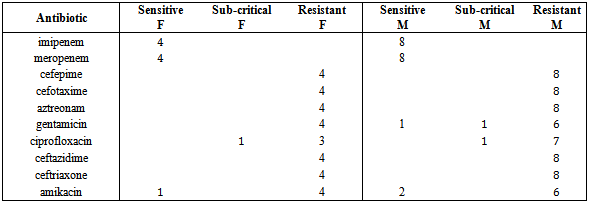 |
| |
|
Table 3. Distribution of patients according to gender
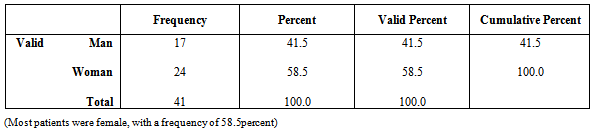 |
| |
|
Table 4. Distribution of the relationship between Resistance gene and patients’ age groups
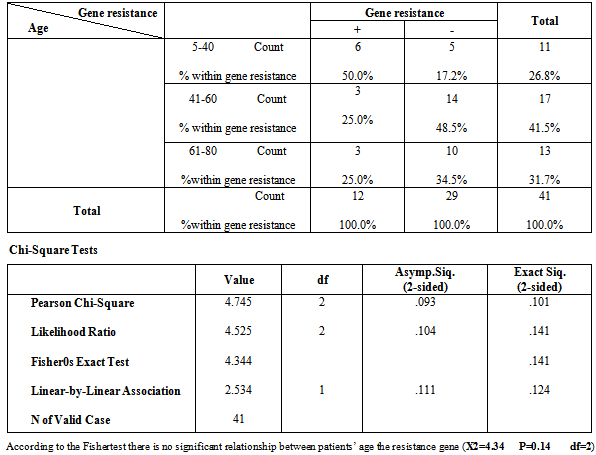 |
| |
|
Table 5. Comparison of the mean age of patients in the resistant gene with non-resistancegenes
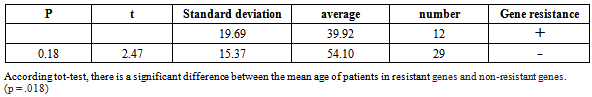 |
| |
|
Table 6. Distribution of the relationship between resistant gene existence and gender
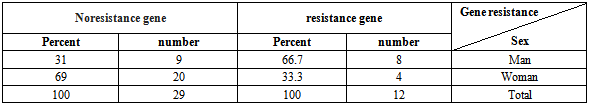 |
| |
|
References
| [1] | Struve, C., Forestier, C. and Krogfelt, K.A. (2003): Application of a novel multi-screening signature-tagged mutagenesis assay for identification of Klebsiella pneumoniae genes essential in colonization and infection. Microbiology, 149, 167–176. |
| [2] | Bailey, R.R.(1987). Urinary Tract Infection. In: Text Book of Renal Disease. Gudith A., White W., editors. New York: Lawrence R Brewer, 196-207. |
| [3] | Mims, C.A., Dockrell, H.M., Goering, R.V. (2004). Medical Microbiology. 3rd edition. London: Mosbey Company. |
| [4] | Leverstein-van, H.M.A., Blok, H.E., Donders, and A.R., et al.(2003): Multi-drugresistance among Enterobacteriaceae is stronglyassociated with the presence ofintegrons and is independent ofspecies or isolate origin. J. Infect. Dis., 187, 251–259. |
| [5] | Schmitz, F.J., Hafner, D., Geisel, and R., et al. (2001): Increasedprevalence of class 1 integrons in Escherichia coli, Klebsiella species, and Enterobacter species isolates over a 7-year period in aGerman university hospital. J. Clin. Microbiol., 39, 3724–3726. |
| [6] | Huaxi, X., Zhaoliang, S., Shengjun, W., et al. (2009): Four novelresistance integron gene-cassette occurrences in bacterial isolates from Zhenjiang, China. Curr. Microbiol., 59, 113–117. |
| [7] | Carattoli, A. (2001): Importance of integrons in the diffusion ofresistance. Vet. Res., 32, 243–259. |
| [8] | Bennet, P.M. (1999): Integrons and gene cassettes: a genetic constructionkit for bacteria. J. Antimicrob. Chemother., 43,1–4. |
| [9] | Radstrom, P., Swedberg, G. and Skold, O. (1991): Genetic analysesof sulfonamide resistance and its dissemination in Gramnegativebacteria illustrate new aspects of R plasmid evolution.Antimicrob. Agents Chemother., 35, 1840–1848. |
| [10] | Leverstein-van, H.M.A., Paauw, A., Box, A.T.A., et al. (2002): Presence of integron-associated resistance in the community iswidespread and contributes to multidrug resistance in thehospital. J. Clin. Microbiol., 40, 3038–3040. |
| [11] | Rao, A.N., Barlow, M., Clark, L.A., et al. (2006): Class 1 integronsin resistant Escherichia coli and Klebsiella spp., UShospitals. Emerg. Infect. Dis., 12, 1011–1016. |


 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML