-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Advances in Life Sciences
p-ISSN: 2163-1387 e-ISSN: 2163-1395
2012; 2(4): 82-84
doi: 10.5923/j.als.20120204.01
The Improved Polymerase Chain Reaction Method Applied for Sex Identification of Crested Ibis, Nipponianippon
Kyung A Kim 1, 2, Jae SeokCha 1, 3, So Young Park 1, 2, Kyung Min Kim 4, Hee Cheon Park 1, 2
1Department of Biology, College of Natural Sciences, Kyungpook National University, Daegu, 702-701, Republic of Korea
2Institute of Ornithology, Kyungpook National University, Daegu 702-701, Korea
3Museum of Natural History, Kyungpook National University, Gunwi, 716-822, Republic of Korea
4School of Applied Biosciences, College of Agriculture and Life Science, Kyungpook National University, Daegu 702-701, Korea
Correspondence to: Hee Cheon Park , Department of Biology, College of Natural Sciences, Kyungpook National University, Daegu, 702-701, Republic of Korea.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
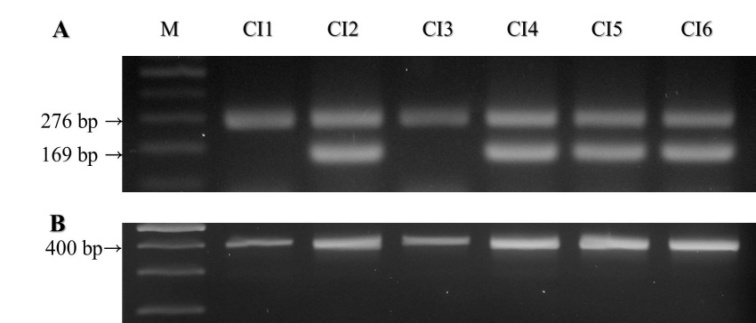
Given the necessity for increased conservation of endangered birds such as the Crested ibis, it is crucial to identify the gender of individuals from species with similar external morphologies among females(ZW) and males(ZZ) to establish a self-sustainable population for restoration of the species. Moreover, raising young chicks during the juvenile period based on sex determination encourages familiarity, which might increase mating success rates. Thus, this study was carried out to determine the sex of Crested ibis chicks using three primers designed to check their identity. A set of polymerase chain reaction(PCR) primers was used to amplify a 276 bp chromo-helicase-DNA binding protein region(CHD) on the Z chromosome in both sexes, whereas the other set was used to amplify a 169 bp female-specific CHD sequence on the W chromosome. The sequences have been submitted to NCBI(GenBank Accession Number: AB620020, AB620021). This method will enable researchers to determine the gender of Crested ibis more simply and quickly.
Keywords: Sexidentification, Crested Ibis, Nipponianippon, CHD, Molecular Sex Identity
1. Introduction
- The Crested ibis, Nipponianippon, is protected as an endangered species by the International Union for Conservation and as a National Treasure in South Korea(No. 198)(1). It is a mediumsized bird that has a long bill and legs, and belongs to the family Threskiornithidae, order Ciconiformes(2). The population has historical ranges in Northeast Asian countries such as Korea, Japan, China, and Far Eastern Russia(3-7), but this range has drastically decreased since the 1930s due to detrimental human activities(8-10). After the last observation of this species in the wild in the early 1980s(11), artificial incubation of this species was conducted for the first time in South Korea(12).For conservation of endangered birds such as the Crested ibis, it is crucial to identify the gender of individuals with similar external morphologies among males(ZZ) and females(ZW) to establish a self-sustainable population(13,14). Moreover, raising young chicks during the juvenile period based on sex determination encourages familiarity, which might increase mating success rates(15). As a result, it is sexes. Therefore, many approaches have been applied to imperative to develop an accurate and way to determine identify sexes currently(16). Compared with other sexidentification approaches, DNA based methods are very effectivebecause a large amount of samples can be tested simultaneously(17).For these reasons, there have been a number of advanced studies regarding the molecular sexing method in this species. As such, although several studies have been conducted to identify the sex of Crestedibis(18-20), the results were unclear or the identification methods were difficult to follow(19). Thus, this study was conducted to design primers to identify the sex of Crestedibis by amplifying Z-specific and W-specific fragments. In addition, due to the 100 bp difference in the sizes of two PCR amplicons, the current method overcomes the difficult situation wherein two fragments are amplified but the size difference between them is too small to allow for sex identification, as in the case of PCR using the P2 and P8 primers. As such, an attempt was made herein to identify the sex of Crestedibis using a single PCR reaction.
2. Materials and Methods
- Sampling and DNA ExtractionThe Crested ibis in this study included a pair of parent Crested ibis(CI1: adult male, CI2: adult female) and their four chicks(CI3, CI4, CI5 and CI6, respectively), which are the first artificially incubated in South Korea. All birds were bred at the Upo Crested ibis Restoration Center in captivity. Tissue specimens were collected from the feathers of the parents and chicks. DNA was extracted from the tissues of the calamus tips using the DNeasy Blood & Tissue Kit(Qiagen GmbH, Hilden, Germany).CHD-W and CHD-Z gene amplification of Crested ibisFirst, PCR of the extracted DNA was performed using the P2 and P8 primers(21). The amplified CHD gene of the Crestedibis was electrophoresed on a 3% agarose gel, subjected to gel extraction and purification, and then sequenced. Using this identified base sequence, primers for sex identification were designed.The forward primer(pci1:5’TGCTGACTTGTACTTTATGTTG3’) and reverse primer(pci3:5’GAGATGGAGTCACTATCAGAT3’), for amplification of the CHD-Z gene of Crested ibis were designed with an expected size for the amplicon of 276 bp. The second forwardprimer(pci2:5’ATGTGGCGTGCCCCCCCTA3’) was designed for specific amplification of the CHD-W gene with an expected amplicon size of 169 bp. As such, sex identification of Crested ibis was performed by PCR using the pci1/2/3 primers. For the PCR, 38 cycles of denaturation(94℃ for 30 sec.), annealing(57℃ for 30 sec.), and extension(72℃ for 60 sec.) were performed with Ex Taq polymerase(TaKaRa, Shiga, Japan) in a DNA Thermal Cycler(TaKaRa TP650, Shiga, Japan). After PCR, the products, the set of pci1, pci2 and pci3 primers, and the other set of P2 and P8 primers, were electrophoresed on a 2% agarose gel.DNA sequencing and alignmentThe amplicons derived from the gel extraction were subjected to cycle sequencing(Applied Biosystems, ABI 3730xl, Foster City, CA, U.S.A.). The final nucleotide sequence data reported in this paper will appear in theDDBJ/EMBL/GenBank nucleotide sequence database with the accession numbers AB620020, AB620021.
3. Results and Discussion
- The PCR products from the male and female were amplified with the pci1, pci2, and pci3 primers.The pci1 and pci2 PCR products were amplified in both the male and female, and the expected size of the amplicon was 276 bp(Figure 1A). In contrast, pci1 and pci3, specifically designed for the identification of sex, produced a product only in the female, and the expected size of the amplicon was 169 bp(Figure 1B). The final nucleotide sequences were submitted toNCBI(GenBank Accession Number AB620020 and AB620021Sex identification thus involves discrimination of the number of fragments present in the PCR reaction: 1 or 2. Therefore, a one-band PCR product indicates a male, and a two-banded PCR product indicates a female.(Figure 2A).PCR was performed using the P2 and P8 primers(Griffiths et al. 1998) and the pci1, pci2 and pci3 primers, after which electrophoresis was performed under the same conditions. Photos of the electrophoresis and the results of the comparison can be seen in Figure 2A and B.Electrophoresis of the amplified products generated using pci1, pci2 and pci3 primers from the adult male(CI1) displayed only a single band while those generated from the adult female(CI2) displayed two bands. Therefore, it was considered that CI3 with one band was male and those with two bands, CI4, CI5, and CI6, were female(Figure 2A).Figure legends
4. Conclusions
- Even when P2 and P8 primers were used in PCR for the identification of sex, which was previously the simplest method as indicated in previous studies, it was still difficult to separate the two electrophoretic bands, although the fragments were amplified successfully. This could be attributed to the small difference between the sizes of the Z- and W-specific fragments(22).Thus, only when electrophoresis was performed in 3% agarose gel for a long time could the two bands be separated(19). In the case of the PCR products created using the pci1, pci2 and pci3 primers, however, separation of the two bands via electrophoresis was easy because the difference between the two bands was 107 bp, which is greater than that between the products created via PCR using the P2 and P8 primers. The set of pci1, pci2 and pci3 primers in our experiment has been thought to be very useful to determine the sex identification in Crested ibis. Therefore this study will promote a molecular genetic study on gender identification of the current Crested ibis.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML