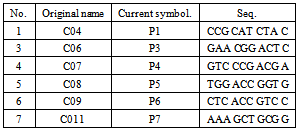
-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Advances in Life Sciences
p-ISSN: 2163-1387 e-ISSN: 2163-1395
2012; 2(2): 9-16
doi: 10.5923/j.als.20120202.03
DNA Fingnerprinting of Eight Cyprinid Fish Species of Iraqi Inland Waters Using RAPD-PCR Technique
Mustafa Sami Faddagh 1, Najah A. Hussain 2, Adnan Issa Al-Badran 2
1Dept. Marine Vertebrate- Marine science Center- Basrah Univ, IRAQ
2Dept. of Biology- College of science- Basrah Univ, IRAQ
Correspondence to: Mustafa Sami Faddagh , Dept. Marine Vertebrate- Marine science Center- Basrah Univ, IRAQ.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
One hundred and twenty specimens of eight cyprinid fish species included Barbus xanthopterus, B. kersin, B. barbulus, B. grypus, B. sharpeyi, B. luteus, Aspius vorax and Cyprinus carpio were collected from Shatt Al-Arab River in governorate of Basrah Southern of Iraq. The DNA fingerprints were identified using RAPD-PCR technique with seven decamere primers. A total of 223 anonymous scorable bands obtained by RAPD-PCR technique across eight cyprinid species genomes. The average of bands per primer was 32 bands, the size of bands estimated using the linear equation of standard curve plot of standard 100 bp ladder. The RAPD markers were used to obtain the similarity indices that were in high value among Barbus species since it ranged from 84.4% between B. xanthopterus and B. kersin to 52% between B. barbulus and B. sharpeyi. While the B. grypus appeared in high similarity indices value (86.9%) with luciscin A. vorax. While C. carpio exhibit high diversity indices value with the Barbus spp.. Therefore the phylogenetic relationships among the eight studied cyprinid species were clearly resolved in dendrograms, they clustered into three branches: the first one included B. xanthopterus, B. kersin, B, barbulus, while the second included B. sharpeyi and B. luteus and the third branch contained B. grypus and A. vorax. Nevertheless, when A. vorax data discarded, the B. grypus return to cluster with B. sharpeyi and B. luteus. Whereas C. carpio functioned as outgroup in both cases. The conclusion of the present study was: molecular patterns showed by RAPD bands represent specific DNA fingerprints and apparently are suitable tool to distinguish among cyprinids. The results clearly revealed that the eight cyprinid taxa are distinctive species. The studied Barbus species are divided into two groups, the first one included B. xanthopterus, B. kersin and B. barbulus which have European Luciobarbus affinities and the second contained B. grypus, B. sharpeyi and B. luteus which they diverged of Luciobarbus but still related to Barbus. So, the Barbus in Iraq is polyphyletic group.
Keywords: RAPD, Fingerprints, Cyprinidae, Iraq, PCR
1. Introduction
- The family Cyprinidae is the most diverse family of freshwater fishes in the world with 220 genera including 2420 species, the family belongs to the order (Nelson, 2006). Whereas Banarescu & Coad (1991) reported more than 2000 species grouped in approximately 340 genera.The role of this family within freshwater ecosystem is central (Calijas and Ochando, 2000) due to including commercial, cultured species and scientific model Danio rario. Barbus is widespread genus, number of species it comprises, the morphological plasticity of species which in some cases make the identification difficult (Callejas and Ochando, 2000), genus Barbus is the most specious in the family Cyprinidae (Karakousis et al., 1995).The major component of Iraqi fish fauna belongs to family Cyprinidae. There are 32 cyprinid species that belong to 16 genera (Coad, 1998; Coad and Hussein, 2007) this represent more than half of total (52 described) species in Iraq while the remaining species belong to other thirteen families.There were few contemporary and recent European systematic studies on Iraqi cyprinids based on morphological characters (Heckle, 1843, Gunther, 1868 ;Berg, 1949).Iraqi fish fauna were described by many local authors, like Khalaf (1961) recorded 45 fish species while Mahdi (1962) described the same number of species as that of the first author. Al-Hamed (1966), Mahdi and Georg (1969), Al-Nasiri and Shamsul-Hoda (1976), Al-Daham (1977) and Al-Rawi et al. (1978). Most of these previous checklists were mainly a compilation of contemporary European authors as noted by Banister (1980) who rejected many records because some of these records were synonyms to others or some of them was not existed in Iraqi fresh waters due to biogeographically factors. The Iraqi fish fauna was recently reviewed by Coad (1991,2009b).One of the main reasons caused the confusion in the description of Iraqi fish fauna was the morphological similarities particularly in the species belong to the same genus and the data compilation without specimens' examination (Banister, 1980).Some of the systematic studies on Iraqi fish fauna were conducted relaying on biometry such as number of fins, fin rays, branched or unbranched, soft or ossified, serrated or unserrated fin rays, number of scales in the lateral line, total and standard length, body width, the percentage of head length to the body lengths etc. (Heckel, 1843 , Gunther, 1868, Berg, 1949, Karaman, 1971; Almaca, 1989) and the number of gill rakers, pharyngeal dentition and some other ostiological characters (Chen et al., 1984). However, such characters are of limited value for identification and differentiation purposes at the genus level, (e.g. Barbus) because this biometrics shows a considerable intraspecific variation while differences among species are small (Callejas and Ochando, 1998). Protein electrophoresis was utilized to discriminate among European Barbus species (Berrebi, et al., 1990; 1995; Machordom et al., 1990; 1995; Karakousis et al., 1995), and of some Iraqi cyprinid including Barbus species (e.g.: Al-Hassan, 1984, 1985; Al-Faisal, 2008). But this technique is unable to distinguish completely among species (Callejas and Ochando, 1998). Whereas Bianco (1998) reported that some Barbus species exhibit limited morphological differences and some external characters are similar, such external characters can be misleading if they depend on environmental conditions. Callejas and Ochando (2000) reported that the morphological and osteologic characteristics are of limited value because they show considerable intraspecific variation and little interspecific differences. And this genus contains species has different levels of ploidy (Berrebi and Tsignopolous, 2003).Currently, researchers started to apply the new technologies based on PCR technique such as recombinant DNA and polymorphic DNA markers and DNA sequence data to resolve questions of fish taxonomy, phylogeny, population genetic and evolutionary biology.Doadrio et al. (2002) used mitochondrial cytochrome b gene sequence to confirm the existence of two previously considered subgenera: Barbus and Luciobarbus and they concluded that there were a closer relationships between subgenus Barbus species with other European Barbus taxa than with Iberian Luciobarbus which has found cluster with North African, Caucasian and Greek species. This result is in agreement with that previously recorded by Bianco (1998) who divided genus Barbus into two subgenera: Barbus and Misinobarbus.There are several Barbus species in Iraqi aquatic ecosystems, like Barbus xanthopterus, B. kersin, B. barbulus, B. grypus, B. sharpeyi, B. luteus which is common, abundant, popular and most commercial (Coad, 2009b).In spite of the puzzle in identification of the first three species (Barbus xanthopterus, B. kersin, B. barbulus), due to the morphological similarity among them but the last three (B. grypus, B. sharpeyi, B. luteus) are in debate till this date since they classified as Tor grypus (Karaman, 1971), Mesopotamichthys sharpeyi (Karaman, 1971) and Carasobarbus luteus (Karaman, 1971). No study was carried out on fish taxonomy in Iraq using molecular markers. The aim of the study is find the finger prints of some barbus species in Iraqi rivers, lakes and wetlands.
2. Materials & Methods
- Sample collectionOne handred and twenty specemns of cyprinids contain Barbus xanthopterus, B. kersin, B. barbulus, B. grypus, B. sharpeyi, B. luteus, A. vorax and Cyprinus carpio were collected from three seperated sites in Shatt Al-arab River in governerate of Basrah contain Qurnah, Garmat ali and Abul-Khaseeb. Fig. 1 is a map showing the collection sites.
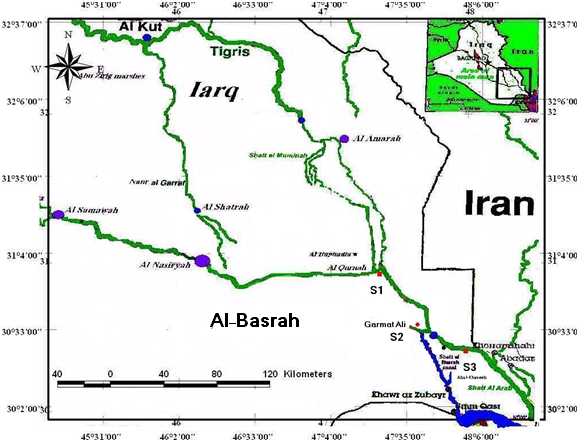 | Figure 1. Map of Shatt Al-Arab River,Basrah in Iraq: Sampling sites: Qurnah (S1),Garmat Ali(S2) and Abul-khasseeb (S3). |
|
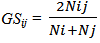 Dendrograms:To show the genetic relatedness among species and populations graphically, the dendrogram were constructed among all species or population tested with RAPD primers. Hierarchical cluster analysis was used of SPSS program (Version 11).
Dendrograms:To show the genetic relatedness among species and populations graphically, the dendrogram were constructed among all species or population tested with RAPD primers. Hierarchical cluster analysis was used of SPSS program (Version 11). 3. Results
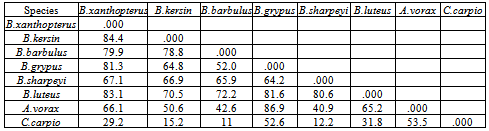
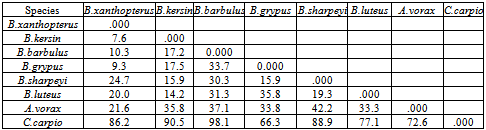
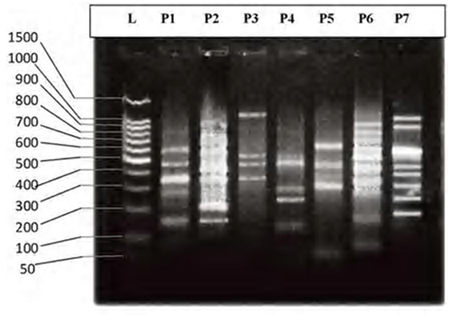
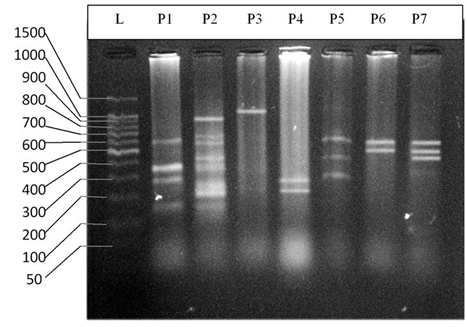
- DNA fingerprinting of Cyprinid species:The number of bands generated per primer varied between 21 and 46 with a mean of nearly 32 bands per primer. The dominant scorable bands were calculated but the weak bands were excluded.Primer P2 and P7 were the most producible primers among the seven RAPD primers used in this study, since the number of produced bands were 46 and 45 respectively while the minimum number (21 bands) produced by P4. In the other hand the primer P2 and P7 were produced the most shared markers among the eight cyprinid fish species which account 15 and 14 bands respectively. Whereas the shared bands yielded by the remaining primers were 29 bands.The molecular profiles obtained from the 2% agarose gel electrophoresis of PCR product documentation for the eight cyprinid species appeared that some of the studied species were totally responded to the seven RAPD primers used in PCR processes, annealed and amplified dominant and recessive bands in their PCR products such as in B. xanthopterus (fig.2), B. kersin (fig. 3), B. barbulus (fig. 4), B. grypus (fig. 5) and Aspius vorax (fig. 8).While few primers did not annealing to some species such as P3 in B. sharpeyi (fig. 6), P1 in B. luteus (fig. 7) and P1and P4 in Cyprinus carpio (fig. 9). The Species varied in the number of bands created by the seven decamere since they ranged from 14 bands in C. carpio to 41 bands in B. xanthopterus.Estimation the size (bp) of amplified bandsIn order to estimate the approximate sizes of bands amplified and scored on the agarose gel electrophoresis, the relationship between the natural logarithm (ln) size of the ladder standard fragments and the distance migrated by that standard fragments had been plotted as standard curve using Windows Office Excel program, there were highly correlated relationships since R2 > 0.97 for all the experiments. The sizes of all bands amplified by the seven primers were calculated by applying the distances migrated by those bands as X and size of ladder standard fragments as Y in the linear equation. The results showed that the seven RAPD primers used to investigate the eight cyprinid species amplified 223 bands ranging from 1444 to 65 bp.The molecular profile of B. xanthopterus using RAPD-PCR with the seven decamere primers exhibited that all the utilized primers amplified a number of bands ranged from three in primer P5 to 10 in primer P6. While the band sizes ranged from 1172 bp in primer P3 to 84 bp in primer P6.The profile of the B. kersin showed that it responded for the seven RAPD decamere primers. The number of amplified bands ranged from single band in primer P3 to eight bands in primer P2. Meanwhile the sizes of amplified bands were from 1172 bp in P3 to 155 bp in P2.In the B. barbulus analysis, the molecular profile also appeared that all the seven RAPD primers generated multiple bands. The number of bands per primer was from two in primer P5 to nine bands in primer P6. The size of bands ranged from 1172 bp in P3 to 122 bp in the same primer.The results showed that also B. grypus responded to all seven RAPD primers used for analysis. The number of amplified bands ranged from single band in primer P6 to six bands in P2 and P7. While the size was from 782 bp in P7 to 116 bp in P2.The molecular profile exhibits that the primer P3 did not anneal to the B. sharpeyi genome. In the same time the remaining six RAPD primers amplified number of bands ranged from single band in P1 to seven bands in P2. While the size of bands was from 1092 bp in P6 to 116 bp in p2.For the B. luteus, the primer P1 did not anneal to the DNA. But the remaining primers generated number of bands from two bands in P4 to five bands in the P3. While the size of bands ranged from 738 bp in P7 to 72 bp in P3.In A. vorax exhibits that it responded to the seven RAPD primers. The bands number per primer ranged from five bands in P2, to seven bands in P7. Whereas the size of bands was from 1444 bp in P7 to 93 bp in P2.In Cyprinus carpio, both P1 and P6 did not anneal with its genomic DNA, but the remaining primers yielded number of bands ranged from six in P7 to two in P2. The size of bands was estimated from 753 bp in P7 to 65 bp in P5.The similarity matrix among the Barbus species were calculated according to band sharing between the studied species but there were apparent differences among them. The similarity indices values (Table 2) ranged from 84.4% between B. xanthopterus and B. kersin to 52% between B. barbulus and B. grypus.In the same time, the remaining similarity values among species were higher than 60%. At all the correlation coefficients among the genetic markers used to calculate the similarity indices among Barbus species were highly supported.In the other hand, the similarity indices values among the eight cyprinid species were ranged from 86.9% between B. grypus and A. vorax to 11% between B. barbulus and C. carpio. While there was noticeable correlation between A. vorax and B. grypus (86.9%).Accordingly, the values of diversity indices among the eight cyprinid species (Table 3) were in low levels among Barbus species, they ranged from 7.6% between B. xanthopterus and B. kerin to 35.8% between B. grypus and B. luteus. But there were high values of diversity indices between Barbus. spp. and C. carpio since the value of diversity indices between B. barbulus and C. carpio was 98.1%.
 | Figure 2. DNA fingerprint of B. xanthopterus migrated on the 2% agarose gel electrophoresis (60V-50min.). L=100 bp ladder and P= primer. |
 | Figure 3. DNA fingerprint of B. kersin migrated on the 2% agarose gel electrophoresis (60V-50min.). L=100 bp ladder and P= primer. |
 | Figure 4. DNA fingerprint of B. barbulus migrated on the 2% agarose gel electrophoresis (60V-50min.). L=100 bp ladder and P= primer. |
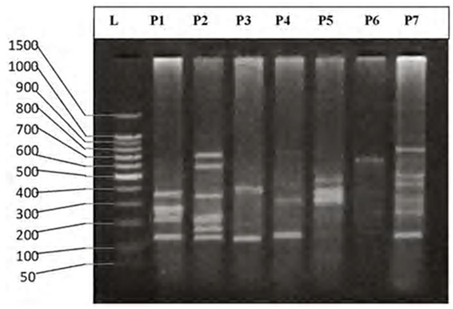 | Figure 5. DNA fingerprint of B. grypus migrated on the 2% agarose gel electrophoresis (60V-50min.). L=100 bp ladder and P= primer. |
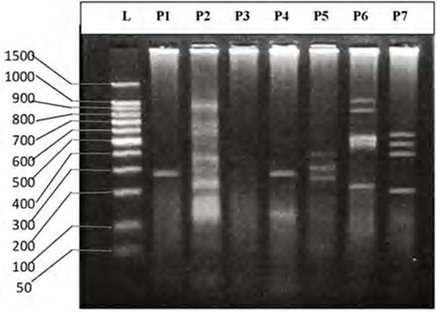 | Figure 6. DNA fingerprint of B.sharpeyi migrated on the 2% agarose gel electrophoresis (60V-50min.). L=100 bp ladder and P= primer. |
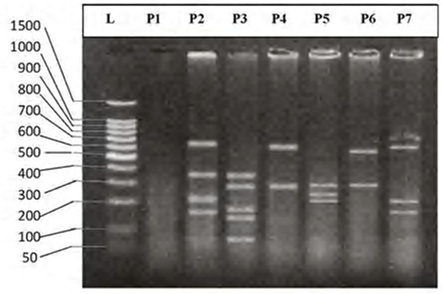 | Figure 7. DNA fingerprint of B.luteus migrated on the 2% agarose gel electrophoresis (60V-50min.). L=100 bp ladder and P= primer. |
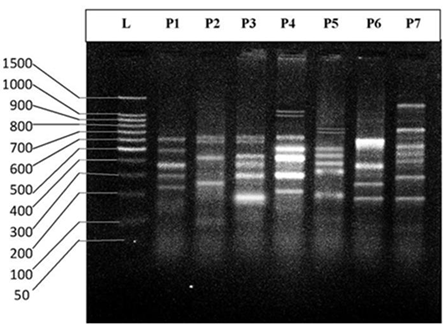 | Figure 8. DNA fingerprint of Aspius vorax migrated on the 2% agarose gel electrophoresis (60V-50min.). L=100 bp ladder and P= primer. |
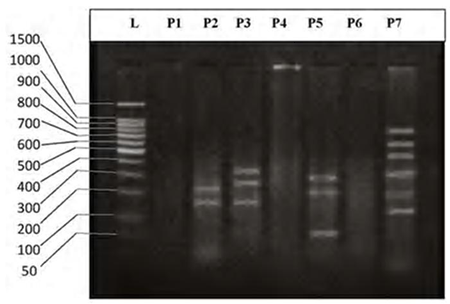 | Figure 9. DNA fingerprint of Cyprinus carpio migrated on the 2% agarose gel electrophoresis (60V-50min.). L=100 bp ladder and P= primer. |
|
|
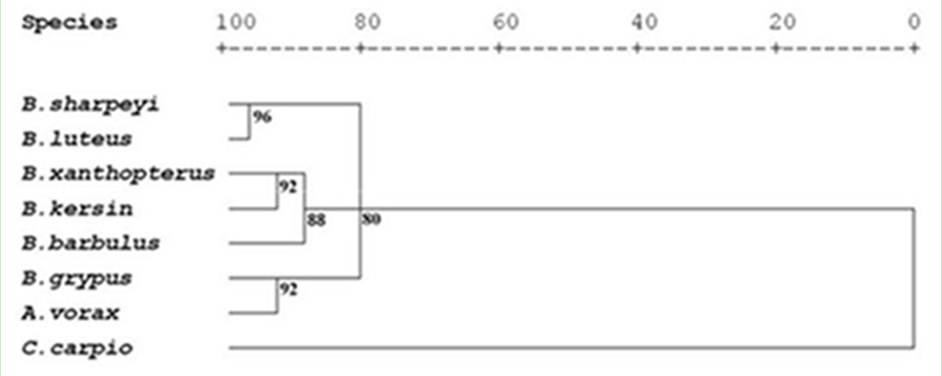 | Figure 10. Dendrogran of eight cyprinu fish species using squared edclidean distance |
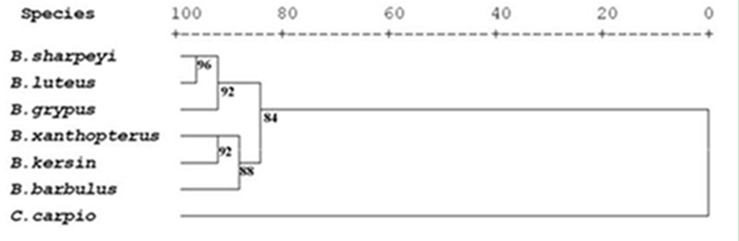 | Figure 11. Dendrogran of six barbus species and cyprinus carpio using squared edclidean distance. |
4. Discussion
- The use of randomly amplified polymorphic DNA (RAPD) protocol to examine genetic variation and to generate DNA fingerprints of some fresh water cyprinids is new to Iraq and paved the way for more future studies to differentiate between fish species existed in Iraqi fresh water and their relation with other in the Middle East and Europe.The randomly amplified polymorphic DNA (RAPD) protocol was reliable, simple to set up, fast and large areas of genomic DNA screened as the study proved that. Consequently this study agrees with earlier studies used the same protocol (Welsh and McClelland, 1990; William et al., 1990). As well as it needs only minute amount of DNA, no prior information about DNA sequence required as that required in the study of mitochondrial and nuclear genes amplification and no hazardous radioactive chemicals used. These advantages make it more preferable than other techniques. The study agree with Gomes et al.(1998) study. The RAPD-PCR amplification with a single decamere primer to produce a DNA fingerprint of DNA fragments was affecting essentially with Primer, DNA template and reaction conditions. So that these advantages in comparison to other DNA fingerprinting method, such as restriction fragment length polymorphism (RFLP) (Fayazi, et al., 2006) and Single Strand conformation polymorphism (SSCP) (Albadran, 2003) that need larger quantity of pure DNA, target gene or few loci of the gene and some information of specific primers or restriction enzymes. In spite of the morphological similarity among cyprinids particularly Barbus species, molecular profiles showed that there was apparent variation in bands amplified by using the same seven RAPD primers. The number of bands on the agarose gel depends on the number of appropriately oriented and target sites present in DNA in that species or individual. This result compatible with the result of Stacey et al.(2007).The results of RAPD profiles showed strongly differentiated fingerprints of the eight cyprinid species, so discrimination among the tested species was easy. This result coincided with the result of Welsh and McClelland (1990) and Hadreys et al. (1992).This procedure established a method to differentiate between cyprinids and other fish families in Iraq. In this context the dendrogram of native Barbus species were dissimilar with the introduced species C. carpio, Two major subgroup were formed among close related species B. xanthopterus ; B. Kersin; B. barbulus and B. sharpyi ;B. luteus; B. grypus(figs 10 &11 ).The present study revealed that the six Barbus species were accurately distinctive species as shown in the RAPD molecular profiles. This result congruent with many morphological and allozyme studies (Al-Hassan, 1984, 1985).Although, Clark and Lanigan (1993) highlighted that the RAPD technique is less laborious compared with other fingerprinting techniques, producing results with low statistical error. In the other hand this method needs accurate work, and multiple decamere primers should be used to generate a number of molecular markers to establish fingerprints. Therefore DNA fingerprint similarity is being used to infer the level of genetic variation within and between natural populations (Lynch, 1990).RAPD variation differed among primers and species since the amplified bands per primer were ranged from 21to 46 and also the bands scored in the eight cyprinid species ranged from 14 to 41. Many studies indicated to those differences (Caccon et al., 1997).RAPD protocol was used to differentiate the cyprinids. This protocol was frequently used to this purpose as example Indian major Carps Labeo rohita, L. calbasu, Catla catla and Cirrhinus mrigala tested to investigate the genetic diversity using RAPD markers (Bardakci and Skibinski, 1994; Barman et al., 2003).For many groups, even qualified taxonomist can’t find characters for reliable identification for early life stages of fish species particularly in cyprinid species due to rapid growth and similar features among them, while using RAPD protocol would solve this problem.RAPD technique is a very useful tool for studying the genetic structure and phylogenetic of the species of Barbus. Also Iberian Barbus species investigated by RAPD to resolve the phylogenetic relationships (Callejas and Ochando, 2001, 2002). The fact that RAPD-PCR technique surveys numerous loci in the genome makes the method particularly attractive for analysis of genetic distance and phylogeny reconstruction (Clark and Lanigan, 1993).RAPD method exhibits more pronounced effect of isolation by distance among populations of Mullus surmuletus in comparison with allozyms (Mamuris et al., 1999).The polymorphism in genetic RAPD markers may stem out of deletion, insertion or substitution in priming sites that cause misleading priming (Williams et al., 1990; Clark and Lanigan, 1993).The additional advantage of RAPD fingerprinting lies in the fact that it is a sequence independent approach and each primer – DNA annealing will produce different spectrum of fragments from the PCR generating a species- specific fingerprint (Rastogi et al. 2007).Actually, the co-migrating dominant markers may not be homologous with one another. Secondly the source of bands use presence/ absence, Although single change in the primer sequence or single substitution, insertion or deletion in gene sequence would alter the product of PCR while scoring that bands as presence/ absence characters causes statistical error (Simmons et al., 2007).
acknowledgements
- This research work is a part of Ph.D. thesis. This research work is a part of Ph.D. thesis. We introduce a gratefulness to all staff in Biotechnology Unit in Science College, University of Basrah.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML