-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Advances in Life Sciences
p-ISSN: 2163-1387 e-ISSN: 2163-1395
2011; 1(1): 1-5
doi: 10.5923/j.als.20110101.01
OlfactionDB: A Database of Olfactory Receptors and Their Ligands
D. Modena 1, M. Trentini 1, M. Corsini 1, A. Bombaci 1, A. Giorgetti 1, 2
1Department of Biotechnology, University of Verona, strada Le Grazie 15, 37134, Verona, Italy
2German Research School for Simulation Science, FZ-Juelich and RWTH Aachen, Germany
Correspondence to: A. Giorgetti , Department of Biotechnology, University of Verona, strada Le Grazie 15, 37134, Verona, Italy.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Odorants are volatile molecules that efficiently carry chemical information, providing one of the main ways for communicating with the environment in all kingdoms of life. In the other hand, mammalian genomes codify for hundreds of olfactory receptors (ORs), e.g. about 400 in human and more than 1000 in mouse, underlying the crucial role of the sense of smell during evolution. Therefore, the olfactory system is capable to discriminate between ~10,000 different odors. The possibility of collecting and compiling information about odorants and their receptors is thus fundamental for a functional characterization of the signaling firing event. OlfactionDB, a manually curated database providing comprehensive information for nearly 400 odorant-receptor interactions at the current state, has been developed for managing information about odorants and their receptors. OlfactionDB is a free publicly database available online from: http://molsim.sci.univr.it/OlfactionDB.
Keywords: Olfactory Receptors, Ligand-Receptor Interactions, Odorant Molecules
Cite this paper: D. Modena , M. Trentini , M. Corsini , A. Bombaci , A. Giorgetti , "OlfactionDB: A Database of Olfactory Receptors and Their Ligands", Advances in Life Sciences, Vol. 1 No. 1, 2011, pp. 1-5. doi: 10.5923/j.als.20110101.01.
1. Introduction
- The G protein-coupled receptors (GPCRs) family is the largest membrane-bound receptor family expressed by mammalians (encompassing more than 1% of the genome). They are involved in an enormous variety of intra- and extracellular signaling pathways, including detection of light, odors and taste; neurotransmission; inflammation; cardiac and smooth muscle contractility[1]. Ligand (or photon) binding to GPCRs fires a cascade of events, producing an electrical signal as output. More than half of the GPCRs codified in mammalian genomes are olfactory receptors (ORs)[2,3], underlying the crucial role of the sense of smell during evolution. With such an impressive number of different ORs, the olfactory system is capable to discriminate between ~10,000 different odors: one odorant can activate numerous types of ORs, while a single OR can be activated by several different odorants. Thus, it is important to rationally collecting and compiling information about interaction affinities of odorants and their receptors. In the last few years, several human/mouse OR-odorants interaction affinities have been published[4-13]. Here we present a free, publicly available, database, OlfactionDB, that contains affinity data of human/mouse olfactory receptors (OR) and ligands, manually compiled and extracted from the literature, containing experimental values for almost 400 OR-odorant interacting pairs.
2. Methods
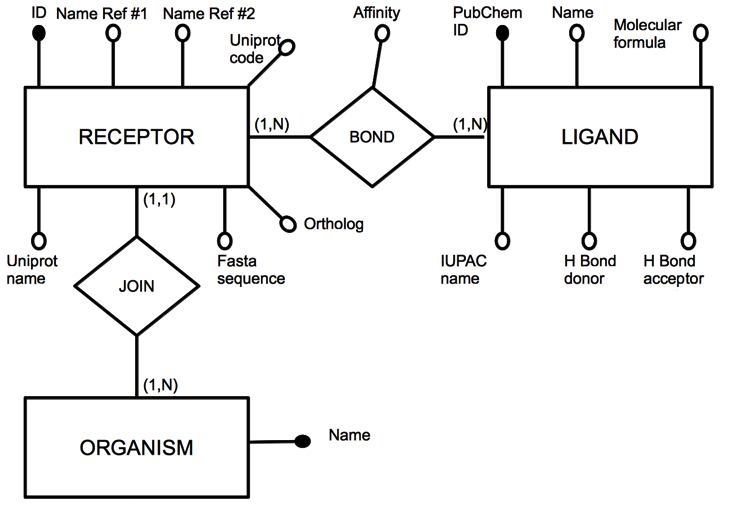
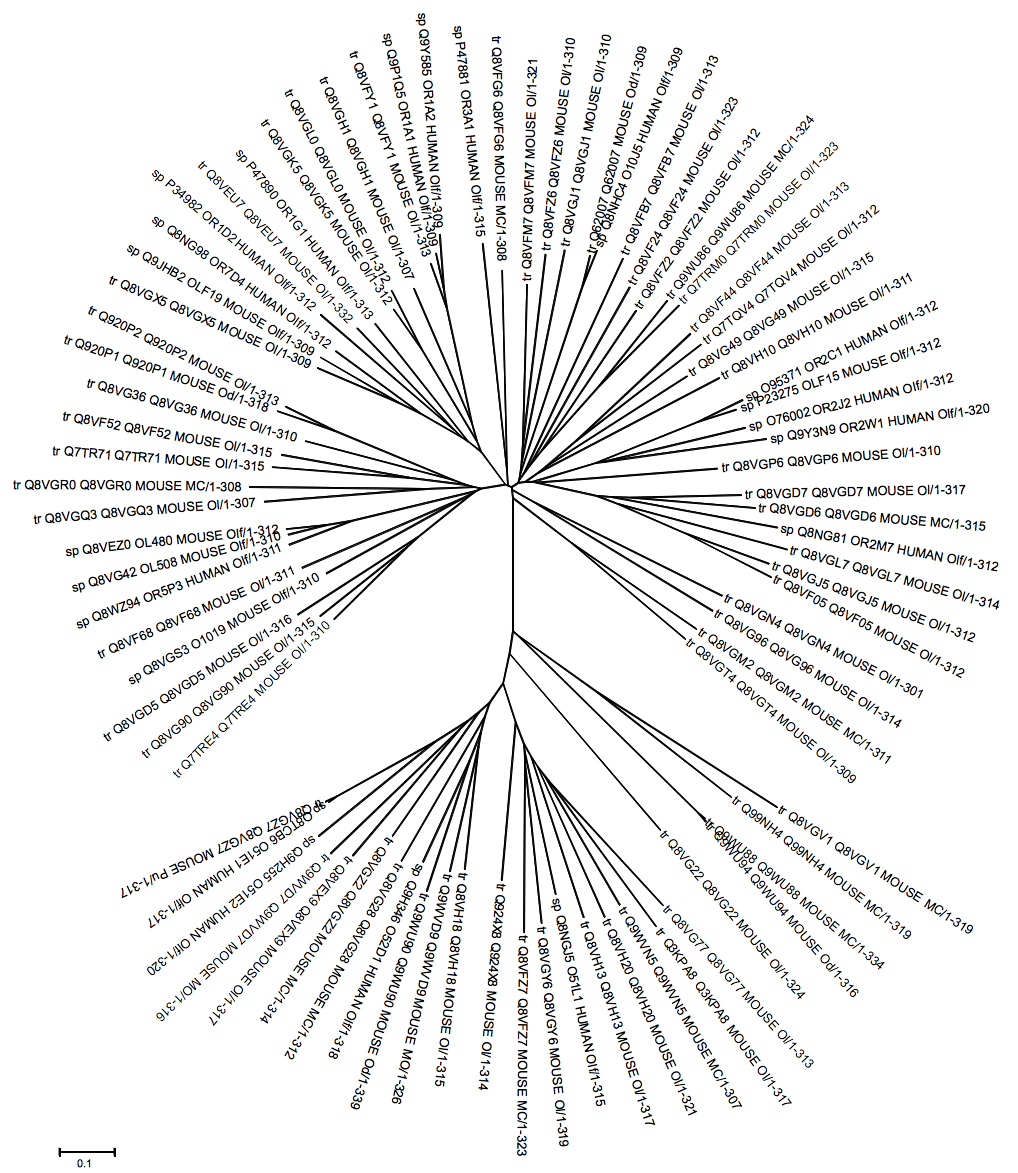
- The whole families of mouse and human ORs were retrieved from the Uniprot database[14]. The ORs for which experimental data exist[4-13] were then aligned using the program PROMALS[15] and manually checked in order to eliminate redundancies. We used in-home-written Python scripts to manipulate the data, whereas annotations regarding the interaction between G-proteins and ligands, the affinity of a particular interaction and the corresponding references were appended manually in a spreadsheet. The data has been organized on the basis of a relational model (Figure. 1) and stored in a PostgreSQL database system. The user has supervisory access through our Apache web server interferential software, which was developed in Java for database manipulation. This software tends to settle any web server’s query. The OR's sequence similarity searches can be carried out in the database using a server-side version of the program Ssearch[16] and the ligand similarity searches can be performed by using a server-side version of OpenBabel (http://openbabel.sf.net).The three dimensional structures of the odorant molecules can be visualized with Jmol[17] and the multiple sequence alignment of the ORs included in the database can be analyzed by a Jalview[18] applet. A tree as the one showed in Figure. 2 can be easily obtained by using the Jalview program.
3. Results
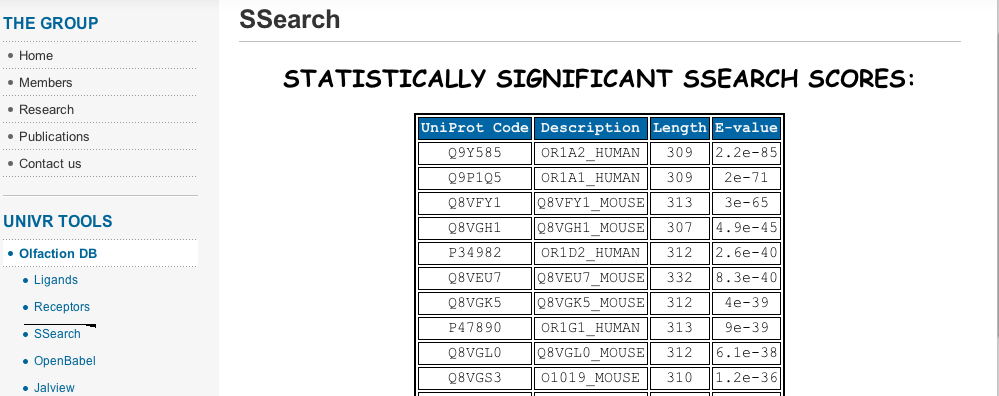
- The current version of OlfactionDB contains comprehensive information for a) 85 odorants, b) 83 odorant receptors, totalizing information for about 400 ligand-receptors pairs. All the data were extracted and annotated from articles published in the last ten years[4-13]. The greatest amount of data was indeed extracted from the seminal work of Saito and co-workers[8].One of the principal problems encountered during the preparation of the database, was the high heterogeneity of nomenclatures found in the different databases explored and on the reference articles. Thus, in order to simplify the database navigation and data retrieval, OlfactionDB offers, through its main page, several search tools: a) full database navigation, b) text and name search, including nomenclatures from at least three different independent sources and c) Uniprot accession code search. Furthermore, similarity searches can be performed using the programs Ssearch and OpenBabel, for receptors and ligands, respectively. An example of a similarity search using Ssearch is shown in Figure. 3.
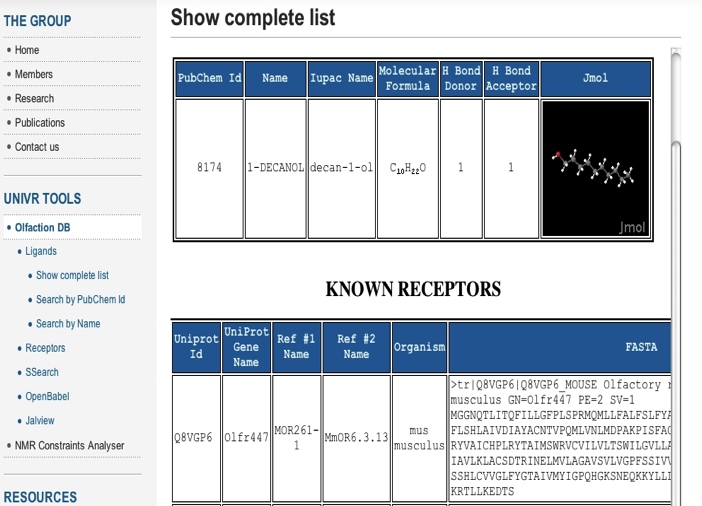 | Figure 4. Typical odorant entry. All the information provided in the webserver is shown: formula, names, amount of H-bonds able to form, structure and its cognate receptors |
4. Discussion
- Olfactory receptors belong to the biggest superfamily of membrane proteins in mammals, i.e. GPCR, and constitute the subfamily with more members. Furthermore, during evolution ORs were shown to have an extremely important role for the survival of complex organisms. A deep insight into the molecular mechanisms underlying the olfactory signalling transduction is thus needed for a complete characterization of the way in which mammals communicate with the rest of the world. The database, i.e. OlfactionDB, here presented can be considered as an initial step into a more profound characterization of the relationships among receptors and their cognate ligands through the whole family of GPCRs. Moreover, detailed information regarding the odorant-receptor affinities may be the key for gaining insights into the structure and the molecular mechanisms underlying the function of the receptors. In particular, this may help in the modelling of the binding cavities, which is the place into which inhibitors and or other kind of molecules may interact. Our group and collaborators have been involved, during the last decade, in the application of a combination of computational and experimental techniques aimed at the unravelling of the molecular mechanisms underlying the different steps of several signalling cascades, i.e. vision, olfaction and bitter taste, all of them including GPCRs as the initial part of the signalling firing event [22-28]. In fact, we have to consider that the main challenges for the near future will include the development and application of methods that permit the full description at the molecular/structural level of GPCR-ligand complexes, as they constitute one of the principal drug targets in the human organism. In this sense the availability of a freely-publicly database, accessible to the whole scientific community, able to offer details regarding ligand-receptor affinities may be of fundamental importance for the development of the field.
ACKNOWLEDGEMENTS
- Funding: Starting Grant from the department of Biotechnology, University of Verona. The authors greatly acknowledge IllyCoffe Company (Trieste, Italy, http://www.illy.com), in particular Dr. Luciano Navarini for very illuminating discussions.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML