| [1] | Monat, J.; Amissah, M.; Gannon, T. Practical Applications of Systems Thinking to Business. Systems 2020, 8, 14. |
| [2] | Monat, J.; Gannon, T. What Is Systems Thinking? A Review of Selected Literature Plus Recommendations. Am. J. of Systems Science 2015, 4, 2. |
| [3] | Monat, J. P.; Gannon, T. Using Systems Thinking to Solve Real-World Problems, College Publications, Rickmansworth, England, U. K., 2017, 2. |
| [4] | Camazine, S.; Deneubourg, J. L.; Franks, N.; Sneyd, J.; Theraula, G.; Bonabeau, E. Self-Organization in Biological Systems, Princeton University Press, Princeton, NJ, USA, 2001, p. 8. |
| [5] | Kosikova, T.; Hassan, N.; Cordes, D.; Slawin, A.; Philp, D. Orthogonal Recognition Processes Drive the Assembly and Replication of a [2]Rotaxane. J. Amer. Chem. Soc. 2017, 137, 16074, https://doi.org/10.1021/jacs.5b09738. |
| [6] | Dieckmann, A.; Beniken, S.; Lorenz, C.; Doltsinis, N.; von Kiedrowski, G. Unravelling a Fulvene Based Replicator: Experiment and Theory in Interplay. Journal of Systems Chemistry. 2010, 1, 10. http://www.jsystchem.com/content/1/1/10. |
| [7] | Liu, Y.; Sumpter, D. Spontaneous Emergence of Self-Replication in Chemical Reaction Systems. arXivLabs. 2018, Cornell University, arXiv:1801.05872v2. |
| [8] | Liu, B.; Pappas, C.; Ottelé, J.; Schaeffer, G.; Jurissek, C.; Pieters, P.; Altay, M.; Marić, I.; Stuart, M.; Otto, S. Spontaneous Emergence of Self-Replicating Molecules Containing Nucleobases and Amino Acids. Journal of the American Chemical Society 2020, 142, 4184-4192, DOI: 10.1021/jacs.9b10796. |
| [9] | Clixby, G.; Twyman, L. Self-Replicating Systems. Org. Biomol. Chem. 2016, 14, 4170, DOI: 10.1039/c6ob00280c, www.rsc.org/obc. |
| [10] | Szostak N.; Wasik S.; Blazewicz, J. Hypercycle. PLoS Comput Biol 2016, 12, e1004853. doi:10.1371/journal.pcbi.1004853. |
| [11] | Vaidya N.; Manapat, M. L.; Chen, I. A.; Xulvi-Brunet, R.; Hayden, E. J.; Lehman, N. Spontaneous Network Formation Among Cooperative RNA Replicators. Nature 2012, 491, 72–7. |
| [12] | Pross, A. Toward a General Theory of Evolution: Extending Darwinian Theory to Inanimate Matter. ACS Nano 2018, 12, 9750–9762; https://doi.org/10.1021/acsnano.8b05821. |
| [13] | Monaco, R.; Montozon, F. (2006). Self-Organization, Autocatalysis and Models of the Origin of Life. Research Gate. 2005, https://www.researchgate.net/publication/228616096, accessed 9/25/2020. |
| [14] | Wood, B. Human Evolution, Oxford University Press, Oxford, England, U.K., 2005, p 16. |
| [15] | New, M.; Pohorille, A. An Inherited Efficiencies Model of Non-Genomic Evolution. Simul. Pr. Theory 2000, 8, 99-108. |
| [16] | Crick, F. Life Itself-Its Origin and Nature, Simon and Schuster; New York, NY, USA, 1981, p 49. |
| [17] | Emmeche, C.; Aspects of Complexity in Life and Science. Philosophica. 1997, 59, 1997, 41-68. |
| [18] | Yeong, F. M. The Complexity of Life: Can Life be Simply Defined? Journal of Biomolecular Structure and Dynamics 2012, 29, 617-618, DOI: 10.1080/073911012010525005. |
| [19] | von Neumann, J. Theory of Self-Reproducing Automata, edited and completed by A. W. Burks. University of Illinois Press, Urbana, IL, USA, 1966. |
| [20] | Bryson, W., A Short History of Nearly Everything, Broadway Books, New York, NY, USA, 2004, p. 185. |
| [21] | DNA Learning Center. RNA Was the First Genetic Molecule. DNA From the Beginning. Cold Springs Harbor Laboratory. http://www.dnaftb.org/26/#:~:text=It%20now%20seems%20certain%20that, is%20easily%20damaged%20by%20enzymes, accessed 2 October 2020. |
| [22] | Parker, N.; Schneegurt, M.; Thi Tu, A.; Forster, B. M.; Lister, P. Microbiology, OpenStax, Rice University, Houston, TX, USA, 2016, p 423. |
| [23] | Hoyle, F.; Wickramasinghe, C. Evolution From Space, Touchstone/Simon & Schuster; New York, NY USA, 1981, pp. 23-24. |
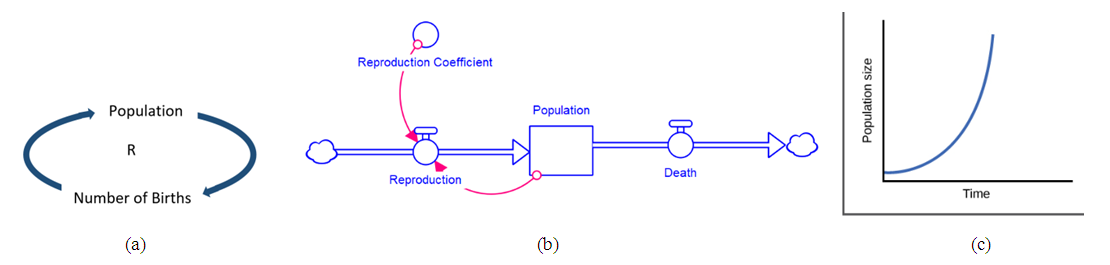
| [24] | Meadows, D., Thinking in Systems, Chelsea Green, White River Junction, VT, 2008, p. 83. |
| [25] | Krissansen-Totton, J.; Arnet, G.; Catling, D. Constraining the Climate and Ocean pH of the Early Earth with a Geological Carbon Cycle Model. Proceedings of the National Academy of Sciences 2018, 115, 4105-4110; DOI: 10.1073/pnas.1721296115 e0bd1681. |
| [26] | Oró, J. Mechanism of Synthesis of Adenine from Hydrogen Cyanide Under Possible Primitive Earth Conditions, Nature 1961, 191, 1193-1194. |
| [27] | Bernal J. D. The Physical Basis of Life. London: Routledge and Kegan Paul: London, England, 1951, p. 364. |
| [28] | Shimoyama, A.; Blair, N.; Ponnamperuma, C. Synthesis of Amino Acids Under Primitive Earth Conditions in the Presence of Clay. In Origin of Life; Noda N. H., Ed., Center for Academic Publisher, Tokyo, Japan, 1978; pp. 95−99. |
| [29] | Yuasa, S. Polymerization of Hydrogen Cyanide and Production of Amino Acids and Nucleic Acid Bases in the Presence of Clay Minerals –In Relation to Clay and the Origin of Life. Nendo Kagaku 1989, 29, 89−96 (in Japanese). |
| [30] | Hashizume, H. Role of Clay Minerals in Chemical Evolution and the Origins of Life. Clay Minerals in Nature – Their Characterization, Modification and Application. 2012. IntechOpen; http://dx.doi.org/10.5772/50172. |
| [31] | Keller, M.; Turchyn, A.; Ralser, M. Non-enzymatic Glycolysis and Pentose Phosphate Pathway-like Reactions in a Plausible Archean Ocean. Mol Syst Biol. 2014, 10, 725; DOI: 10.1002/msb.20145228. |
| [32] | Todisco, M.; Tommaso, P.; Fraccia, T. P.; Smith, G. P.; Corno, A.; Bethge L.; Klussmann, S.; Paraboschi, E. M.; Asselta, R.; Colombo, D.; Zanchetta, G.; Clark, N. A.; Bellini, T. Nonenzymatic Polymerization into Long Linear RNA Templated by Liquid Crystal Self-Assembly, ACS Nano 2018, 12, 9750–9762. |
| [33] | Powner, M. W.; Gerland, B.; Sutherland, J. D. Synthesis of Activated Pyrimidine Ribonucleotides in Prebiotically Plausible Conditions, Nature 2009, 459; doi:10.1038/nature08013. |
| [34] | Orgel, L. E. Prebiotic Chemistry and the Origin of the RNA World. Crit. Rev. Biochem. 2004, 39, 99-123; doi: 10.1080/10409230490460765. |
| [35] | Israel, B. New Study Brings Scientists Closer to the Origin of RNA. Georgia Tech News Center. 2013. http://www.news.gatech.edu/2013/12/23/new-study-brings-scientists-closer-origin-rna, accessed 10/2/2020. |
| [36] | Chen, M.; Cafferty, B.; Mamajanov,I.; Gállego, I.; Khanam, J.; Krishnamurthy, R.; Hud, N. Spontaneous Prebiotic Formation of a β-‐ribofuranoside that Self-Assembles with a Complementary Heterocycle. J. Am. Chem. Soc. 2014, 136, 5640–5646. |
| [37] | Becker, S.; Thoma, I.; Deutsch, A.; Gehrke, T.; Mayer, P.; Zipse, H.; Carell, T. A High-yielding, Strictly Regioselective Prebiotic Purine Nucleoside Formation Pathway. Science. 2016, 352, 833-6; DOI: 10.1126/science.aad2808. |
| [38] | Szostak, J. W. Systems Chemistry on Early Earth, Nature 2009, 459, 172. |
| [39] | Sanchez, R. A.; Ferris, J. P.; Orgel, L. E. Cyanoacetylene in Prebiotic Synthesis, Science 1966, 154, 784–785. |
| [40] | Thaddeus, P. The Prebiotic Molecules Observed in the Interstellar Gas. Phil. Trans. R. Soc. B 2006, 361, 1681–1687. |
| [41] | Lincoln, T.; Joyce, G. Self-Sustained Replication of an RNA Enzyme. Science. 2009, 323, 1229–1232; DOI:10.1126/science.1167856. |
| [42] | Robertson, M. P.; Joyce, G. F. Highly-Efficient Self-Replicating RNA Enzymes. Chem Biol. 2014, 21, 238–245; doi:10.1016/j.chembiol.2013.12.004. |
| [43] | Cojocaru, R.; Unrau, P. Origin of life: Transitioning to DNA genomes in an RNA World. Insight. 2017, eLife 2017; 6: e32330 DOI: 10.7554/eLife.32330. |
| [44] | Lazcano A,; Fastag, J.; Gariglio, P.; Ramírez, C.; Oró, J. The Evolutionary Transition from RNA to DNA in Early Cells. Journal of Molecular Evolution. 1988, 27, 283–290. |
| [45] | Meselson, M.; Stahl, F. The Replication of DNA in Escherichia coli. PNAS. 1958, 44, 671–82. DOI: 10.1073/pnas.44.7.671. PMC 528642. PMID 16590258. |
| [46] | Kishimoto, T. Entry into Mitosis: a Solution to the Decades-long Enigma of MPF. Chromosoma. 2015. 124, 417–428; DOI 10.1007/s00412-015-0508-y. |
| [47] | Lindquist, A.; Rodriguez-Bravo, V.; Medema, R. The Decision to Enter Mitosis: Feedback and Redundancy in the Mitotic Entry Network. J. Cell. Biol. 2009, 185, 193-202. |
| [48] | Nishitani, H.; Lygerou, Z.; Control of DNA Replication Licensing in a Cell Cycle. Genes to Cells 2002, 7, 523-534. |
| [49] | Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific Enzymatic Amplification of DNA in Vitro: The Polymerase Chain Reaction. Cold Spring Harbor Symposia on Quantitative Biology 1986, 51, 263–273. DOI: 10.1101/sqb.1986.051.01.032. PMID 3472723. |
| [50] | Moelling, K.; Broecker, F. Viruses and Evolution – Viruses First? A Personal Perspective. Mol. Biol. 2019, 39, 99–123, DOI.org/10.3389/fmicb.2019.00523. |
| [51] | Khan Academy. Evidence for Evolution. https://4m.cn/sxqZO, accessed 2 October 2020. |
| [52] | Tamura, K.; Subramanian, S.; Kumar, S. Temporal Patterns of Fruit Fly (Drosophila) Evolution Revealed by Mutation Clocks, Mol. Biol. Evol. 2004, 21, 36–44; DOI: 10.1093/molbev/msg236. |
| [53] | Crick, F. Life Itself-Its Origin and Nature, Simon and Schuster; New York, NY, USA, 1981, pp. 171-175. |
| [54] | Deamer, D.; Singaram, S.; Rajamani, S.; Kompanichenko, V.; Guggenheim, S. Self-Assembly Processes in the Prebiotic Environment. Philos Trans R Soc Lond B Biol Sci. 2006, 361, 1809-18; DOI: 10.1098/rstb.2006.1905. |
| [55] | Fox, S.; Harada, K. Thermal Copolymerization of Amino Acids to a Product Resembling Protein. Science. 1958, 128, 1214; DOI: 10.1126/science.128.3333.1214. |
| [56] | Rohlfing, D. L.; McAlhaney, W. W. The Thermal Polymerization of Amino Acids in the Presence of Sand, Biosystems, 1976, 8, 139-145; https://doi.org/10.1016/0303-2647(76)90016-2. |
| [57] | Leman, L.; Orgel, L.; Ghadiri, M. Carbonyl Sulfide-Mediated Prebiotic Formation of Peptides. Science. 2004, 306, 283-6; DOI: 10.1126/science.1102722. |



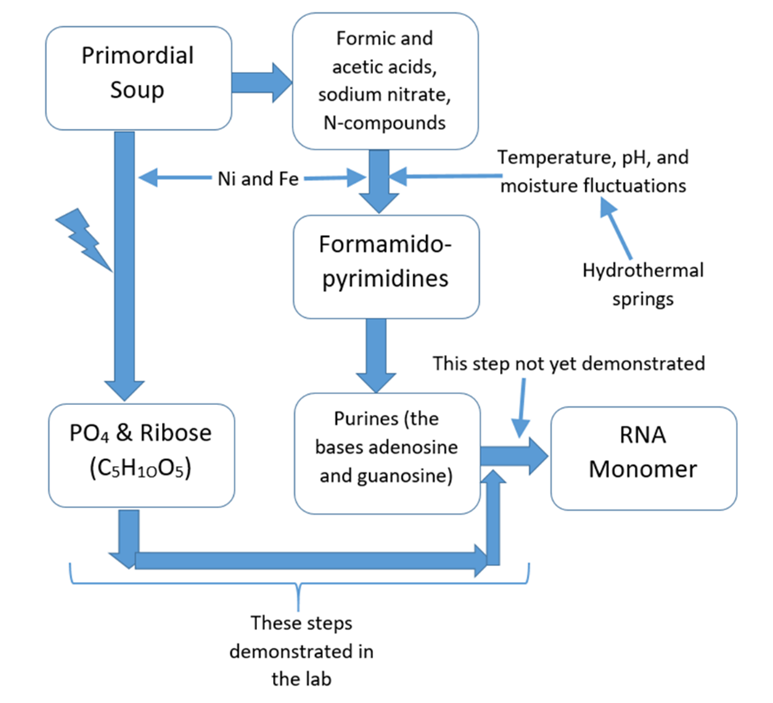
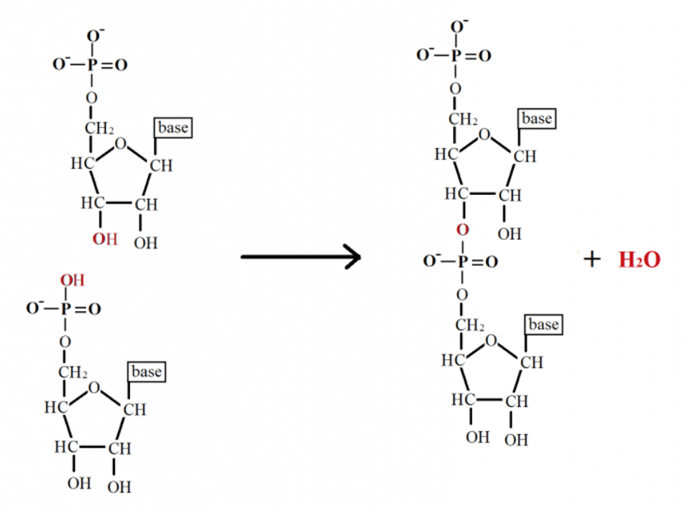
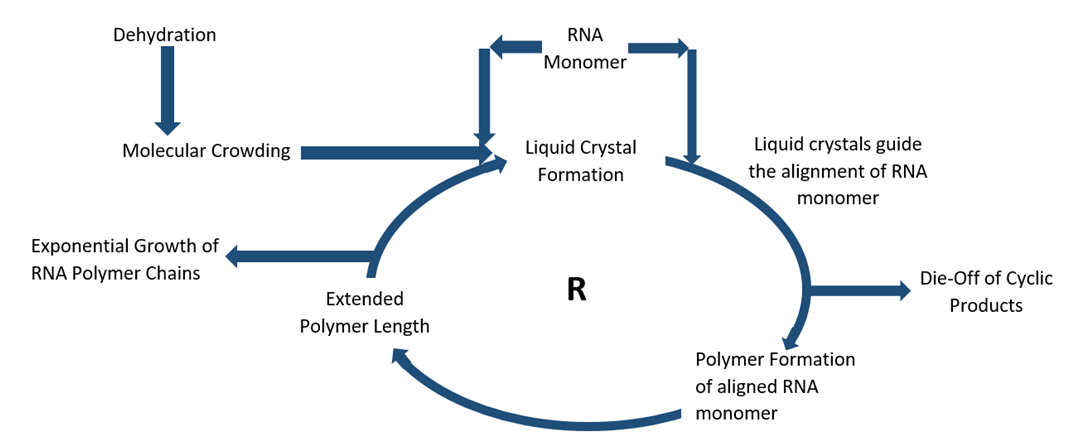
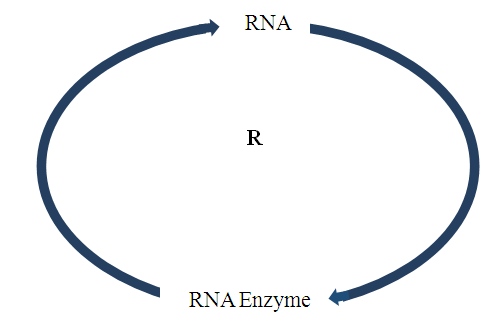
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML