Kadirov J. F.1, Rizaev J. A.1, Ziyadullaev Sh. Kh.2, Boboev K. T.3, Fayzullaeva D. B.4, Shodieva D. A.1, Dushanova G. A.5
1Samarkand State Medical University, Samarkand, Uzbekistan
2Institute of Immunology and Human Genomics, Academy of Sciences of the Republic of Uzbekistan, Tashkent, Uzbekistan
3Republican Specialized Scientific and Practical Medical Center of Hematology, Tashkent, Uzbekistan
4Republican AIDS Center, Tashkent, Uzbekistan
5Samarkand State University, Samarkand, Uzbekistan
Copyright © 2025 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Abstract
This study examined the associations between the HLA-DRA gene polymorphism rs8084 and the expression of key antiviral genes MX2, IFNM1, and ADAR1 in HIV-1-infected patients of the Uzbek population in the Samarkand region. The results revealed significant differences in the frequency of alleles and genotypes of the HLA-DRA gene between the control and patient groups. The T allele was associated with an increased risk of HIV development, while the C allele, on the other hand, may provide some protection. Notably, it was found that the genotypes C/C and C/T, exhibiting increased expression of MX2, IFNM1, and ADAR1 genes in HIV-infected individuals, could play an important role in protection against the virus. Specifically, patients with the T/T genotype demonstrated significantly lower levels of gene expression, indicating a potential weakening of the antiviral immune response.
Keywords:
Polymorphism, Heterozygote, Genotype, MX2, IFNM1, ADAR1, HIV-1
Cite this paper: Kadirov J. F., Rizaev J. A., Ziyadullaev Sh. Kh., Boboev K. T., Fayzullaeva D. B., Shodieva D. A., Dushanova G. A., Interrelationship Between the Rs8084 Polymorphism of HLA-DRA C and the Expression of the MX2, IFNM1, and ADAR1 Genes in HIV-1 Infected Patients, American Journal of Medicine and Medical Sciences, Vol. 15 No. 3, 2025, pp. 599-605. doi: 10.5923/j.ajmms.20251503.24.
1. Introduction
The relationship of HLA-DRA genotype polymorphism with the expression of MX2, IFNM1, and ADAR1 genes is an important topic in the field of molecular immunology and genetics. The HLA-DRA gene encodes an alpha chain that is part гистосовместимостиof the MHC Class II major histocompatibility complex and plays a key role in the immune response, as they are involved in the presentation of antigens to T cells, which is an important process for activating immune cells and creating a specific immune response. Polymorphism in the HLA-DRA gene can affect the body's ability to effectively recognize and respond to pathogens or altered cells, which can play an important role in various diseases and immune responses [1,2]. Polymorphism in the HLA-DRA gene can affect the expression of the MX2, IFNM1, and ADAR1 genes through several mechanisms. As aninfluence on interferon activation, the HLA-DRA gene plays an important role in T cell activation and can influence interferon activation in response to antigenic stimuli. Since MX2 and IFNM1 are actively regulated by interferons, polymorphism in HLA-DRA can affect the intensity of their expression. ОRestricted HLA-DRA alleles can increase or decrease the activity of the interferon response, which in turn affects the expression of MX2 and IFNM1 [3]. The MX2 Myxovirus Resistance 2 gene encodes a protein that plays an important role in protecting the body from viral infections. The MX2 protein is anti-replicative, preventing the replication of viruses such as HIV, influenza, and others. MX2 expression is enhanced in response to interferon activation, which makes it an important element in the body's immune response to viral infections [5].Interferon production activates antigen-presenting cells, such as macrophages and dendritic cells, and also stimulates the expression of antiviral genes, such as MX2 [4]. Polymorphism in HLA-DRA can affect the efficiency of antigenic presentation and the rate of activation of antiviral activity, which in turn can modulate the severity of MX2 expression. IFNM1 Interferon, Alpha-inducible protein 1 is a gene that encodes a protein activated by interferons and is involved in the response to viral infections. Interferons are important molecules that activate the body's antiviral response. They increase the expression of a number of genes, including MX2, that play a role in fighting viral infection.ADAR1 Adenosine Deaminase Acting on RNA 1 is a gene that encodes an enzyme responsible for RNA editing, in particular for the deamination of adenosine to inosine. This process plays an important role in the regulation of gene expression, RNA stabilization, and immune response to viral infections. ADAR1 is involved in the modification of viral RNA and can help modulate the immune response, preventing excessive activation of the immune system [7].ADAR1 plays an important role in regulating immune responses and stabilizing RNA, especially in the context of viral infections. Some polymorphisms in HLA-DRA can affect ADAR1 activity through changes in the level of interferons or other molecules, which can change the processing of RNA by viruses or cells that trigger an immune response. Polymorphisms in the HLA-DRA genes may also affect ADAR1 expression, as these two genes may be linked in regulating the cellular response. HLA-DR may be involved in the activation or inhibition of ADAR1 expression via signaling pathways associated with the immune response [5,7].
2. Material and Methodology
DNA samples of men and women of Uzbek nationality were used for the study. 83 patients with HIV diagnosis aged 14 to 66 years were examined. The control group consisted of 63 practically healthy individuals aged 15 to 57 years, unrelated, without clinical signs of HIV infection and hereditary burden of this disease. All participants in the study signed an informed consent form. The sex ratio in the examined group of HIV-1 infected people was 1: 1 (n=43 men, n =40 women). The molecular and genetic part of the work was performed at the RSPMC of Hematology of the Ministry of Health of the Republic of Uzbekistan, in the Department of Molecular Medicine and Cell Technologies. The level of expression of the ADAR1, MX2, and IFTM1 genes was studied by quantitative PCR using the " Kits for assessing the expression of ADAR1, MX2, and IFTM1 genes"from Inogene (Russia).Genotyping of polymorphic loci consisted of several stages:1. Peripheral blood sampling;2. Isolation of mRNA from lymphocytes;3. cDNA synthesis from mRNA;4. Detection of polymorphisms by quantitative - PCR;Comparative analysis of the following genes: ADAR1, MX2, IFTM1, was performed using a "case-control" model comparing two comparative groups or subgroups. The case sampleconsisted» состояла of 83 patients with HIV1 infection. The mean age of these patients was (32±7.8). The control sample consisted of 63healthy donors of Uzbek nationality who corresponded by sex and age to the examined group of patients (p>0.05), without any concomitant pathologies. The search for ADAR1, MX2, and IFTM gene sequences for the selection of oligoprimes was performed in NCBI GenBank (http://www.ncbi.nlm.nih.gov/GenBank). The nucleotide sequences and oligoprimer characteristics of these genes were evaluated using the исползованием пакет программы «Oligo v. 6.31 software package (Molecular Biology Insights Inc., USA).Each reaction mixture with a volume of 25 µl during PCR included a mixture of deoxynucleotide triphosphates 10 M 0.5 µl, 1 e. a. TaqDNA polymerase, 10 x Gold Star buffer 0.5 µl, MgCl210 mm 1.5 µl, 0.1 µm of each primer, and 1 µl of genomic DNA at a concentration of 0.1 µg / ml.Biomaterial sampling was performed using standard vacuum tubes containing EDTA-K3 anticoagulant (Vacutainer Becton Dickinson International, USA). mRNA isolation for PCR studies was isolated from 100 µl of venous blood using the TriZ комплектreagent kit «TriZ reagent (Inogene, Russia). mRNA concentrations were measured on a NanoDrop 2000 spectrophotometer (NanoDropTechnologies, USA) at A260/280 nm wavelength. The purity of all mRNA samples was 1.7/1.8. Detection of ADAR1, MX2, and IFTM 1 gene accumulation products1, was performed by real-time quantitative PCR.Синтез cDNA synthesis was performed using the commercial kit" Reverzyme" (Inogen, Russia), using термоциклера a CFX96 thermal cycler (Biorad, USA) in accordance with the following amplification programs:Incubation - 65°S (5 minutes), 37°S (37 minutes), хранение storage.PCR analysis was performed in "real-time" mode with fluorescent detection of accumulation of DNA amplification products using термоциклеров Rotor Gene Q thermocyclers (Corbett Research» QUAGEN Germany) and CFX96 (Biorad, USA), and in accordance with the following amplification programs:Pre-denaturation – 95°S (10 min), 1 amplification cycle, 95°S (10 sec – - denaturation, 60°S (50 sec) - primer annealing, and final synthesis. 47 amplification cycle, 10 min storage at 4°S.In accordance with the manufacturer's instructions, each gene was determined based on the intensity and combination of bands of amplified fragments in the "target gene"and" normalizer gene".The results of testing and detection of polymorphisms of the following genes: ADAR1, MX2, IFTM1 in the group of patients n=83 and in the control group n=63 are presented in Figures 1 and 2.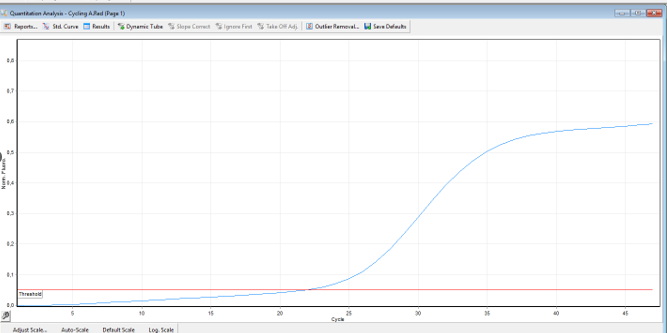 | Figure 1. The result of real-time PCR (detection of the target gene) |
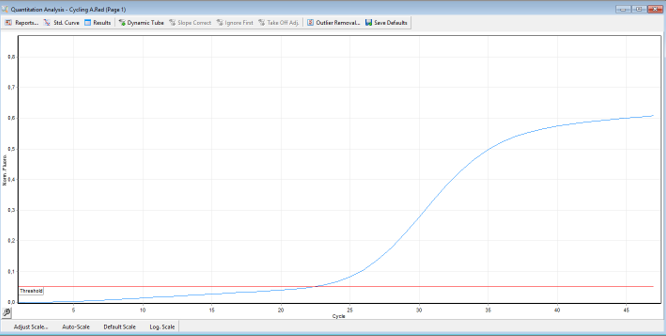 | Figure 2. The result of real-time PCR (detection of the GUSB normalizer gene) |
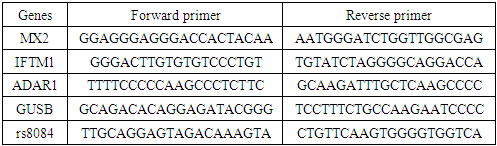
The following primer sequences were used for real-time PCR analysisTable 1. Primers of the studied genes
 |
| |
|
Statistical data processing was performed using the programs Statistica for Windows v. 6.0, MS Excel ' 98 (Microsoft) [14]. The mean values, standard deviations, standard errors of the mean, and 95% confidence intervals were calculated. Genotype and allele frequencies were compared using the exact two-way Fisher test (P). The Bonferroni correction (P') was used to correct for multiple comparisons. The relative risk of the disease (OR) was calculated using the formula of J. M. Bland (2000) [15].
3. Research Results
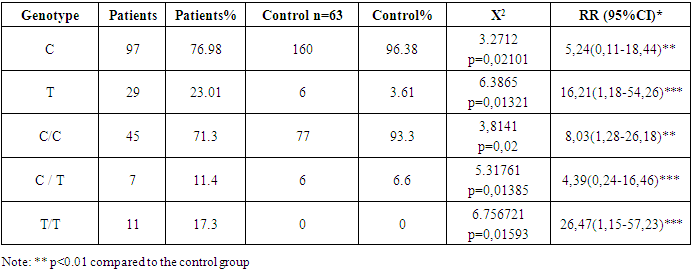
In case-control studies, 83 HIV-infected patients and healthy individuals were recruited sequentially. Demographic data and clinical characteristics of the study groups are summarized in Table 1. Significant differences were observed by age and gender between the case and control groups (P<0.001 and P<0.001), respectively.Table 2. Frequency distributions of alleles and genotypes of the HLA DRA, rs 8084 gene in the Uzbek population of the Samarkand region
 |
| |
|
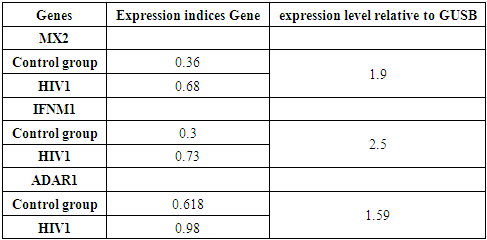
The study of the frequency distribution of alleles and genotypes of the HLA DRA gene, rs8084 polymorphism in the population of the Samarkand region showed significant differences between the control group and the group of HIV patients. Analysis of the frequency of distribution of the C allele, the frequency аллеляof the C allele in the control group was 96.38%, which is significantly higher than in the group of HIV patients, where the frequency of this allele is 76.98% (p=0.02101). The odds ratio (RR) for аллеляthe C allele in the group of HIV patients was 5.24 (95% CI: 0.11-18.44), which indicates an increased risk of HIV infection in carriers of this allele. The frequency of the T allele was studied, the frequency of the T allele in the control group was 3.61%, and in the group of HIV patients 23.01% (p=0.01321). The odds ratio (RR) for аллеляthe T allele in the group of HIV patients was 16.21 (95% CI: 1.18-54.26), which indicates a significant increase in the risk of developing HIV infection in carriers of this allele.Individuals with the C/C genotype had a C/C genotype prevalence of 93.3% in the control group and 71.3% in the HIV group (p=0.02). The odds ratio (RR) for the C/C genotype in the group of HIV patients was 8.03 (95% CI: 1.28-26.18), which confirms a lower frequency of this genotype among patients, which may indicate a protective effect of homozygous carriage аллеляof the C allele. Individuals with the C/T genotype, the frequency of the C/T genotype in the control group It was 6.6% in this group and 17.02% in the HIV group (p=0.01385). The odds ratio (RR) for the C/T genotype in the group of HIV patients was 4.39 (95% CI: 0.24-16.46), which indicates a moderately increased risk of HIV infection among carriers of this genotype, although the statistical significance of these data is less pronounced.T/T genotype, T/T genotype was found only in the group of HIV patients, while it was not found in the control group (p=0.01593). The odds ratio (RR) for the T/T genotype was 26.47 (95% CI: 1.15-57.23), which indicates a high risk of HIV infection in carriers of this genotype.Thus, the polymorphism of the HLA DRA gene (rs8084) has a significant impact on the predisposition to HIV infection in the population of the Samarkand region. Carriers of the T allele and T/T genotype, in particular, have a significantly higher risk of developing HIV compared to carriers of the C allele and C/C genotype. These findings highlight the possible role of the HLA DRA gene in the immune response to HIV infection and can be used for further research to develop personalized HIV prevention and treatment methods. Overall, the results of this study confirm the importance of studying the HLA DRA gene polymorphism as a potential marker for assessing the risk of HIV infection in different ethnic groups. The presence of significant associations between different HLA-DRA rs8084 genotypes and the studied traits/diseases in the Uzbek population of the Samarkand region may indicate a genetic predisposition to certain conditions, such as immune or inflammatory diseases. ГомозиготаThe C/C and T/T homozygotes, as well as certain heterozygotes such as C/T, show a high risk, which requires further research to establish the specific mechanisms underlying these associations.This analysis can be useful for understanding genetic predisposition to diseases and for developing personalized approaches to diagnosis and treatment based on genetic markers.Theезультаты экспрессии генов results of expression of the MX2, IFNM1, and ADAR1 genes in patients with HIV1 infection are presented in Table 3. For each of the genes, expression indicators are shown for both the control group and the group of HIV1 patients. Theровень экспрессии level of expression of the MX2 gene MXin relation to the control GUSB gene, which is used for data normalization, shows the level of expression of the MX2 gene in the control groupило, or 0.36, which corresponds to a value of 1.9 in relation to the GUSB gene.Table 3. Results of expression MXof MX2, IFNM1,and ADAR1 genes in patients with HIV-1 infection
 |
| |
|
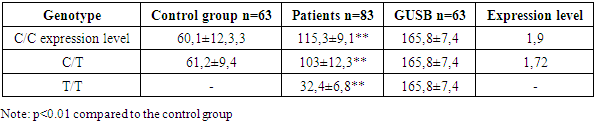
In patients with HIV1, the expression level of the MX2 gene is increased to 0.68, which also indicates an increase in the activity of this gene in the group of infected people.The level of expression of the IFNM1 gene is 0.3, which gives a value of 2.5 relative to the GUSB gene; у in patients with HIV1, the level of expression of the IFNM1 gene is increased to 0.73, which indicates greater activation of this gene in infected patients compared to the control group.In the control group, the expression level of the ADAR1 gene is 0.618, which corresponds to a value of 1.59 relative to the GUSB gene;, у in patients with HIV1, the expression level of the ADAR1 gene is also increased and is 0.98, which indicates an increased activity of this gene among infected people.Thus, the table shows that in patients with HIV1, the expression of the MX2, IFNM1 and ADAR1 genes is increased compared to the control group, which may reflect an enhanced immune response of the body to the virus.HLA-DRA of the MHC II molecule, which plays a key role in the immune system by presenting antigens to T cells, pterymorphisms in this gene can affect the body's ability to recognize and respond to pathogens, including viruses such as HIV. HLA-DRA can alter the efficiency of viral antigen presentation, which affects the immune response against HIV. In particular, alleles and genotypes that increase or decrease HLA-DRA expression can modulate T cell activity and thereby alter the clinical outcome of HIV infection, and alsoазываетaffect interferon activity, which in turn activates MX2. Table 4 shows data on the level of expression of the MX2 gene in HIV1-infected patients, depending on the genotype of the rs8084 HLA-DRA polymorphism. Also, Table 4 shows indicators for the control group and for the group of individuals in relation to the control GUSB gene, the level of expression of the GUSB gene, which is used as a control for data normalization. Table 4. Expression parameters of MX2 genesin HIV1 infected patients depending on the genotype of the rs8084 HLA-DRA polymorphism
 |
| |
|
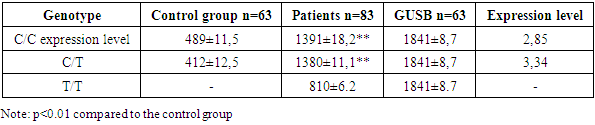
In the presented genotypes of the rs8084 HLA-DRA polymorphism C/C, C/T and T/T, the level of expression of the MX2 gene in healthy individuals of the control group the level of expression of the MX2 gene in comparison with HIV-infectedindividuals was revealed, уin individuals with the C/C genotype of the rs 8084 polymorphism of the HLA-DRAgene, in the control group there were values of 60.1 ± 12.3, у бin mostindividuals 115.3 ± 9.1 (p≤0.01) in relation to the GUSB gene 165.8 ± 7.4, There were 1.9 times higher values of the MX2 gene expression in the group of patients with HIV1 infected individuals compared with the control group.Individuals with the C/T genotype of the rs 8084 polymorphism of the HLA-DRA, gene showed a 1.72-fold higher level of MX2 gene expression in individuals infected with HIV1 compared to the control group (61,2±9,4, 103±12,3, GUSB: 165.8±7.4, p≤0.01).Individuals with a homozygous T/T genotype of the rs 8084 polymorphism of the HLA-DRAgene were not detected in the control group of this genotype, the results in HIV1 infected patients show lower expression rates of the MX2 gene 0.52 times (32.4±6.8, GUSB: 165.8±7.4, p≤0.01) compared to the C/C and C genotypes./T. The highest level of expression of the MX2 gene is observed in patients with the C/C genotype 1.9, followed by the C/T genotype 1.72. In patients with the T/T genotype, the level of MX2 expression is significantly reduced, which may indicate a possible link between this genotype and a lower level of MX2 expression in HIV-infected individuals.Thus, allele C may be associated with a higher expression of HLA-DRA, which leads to an increased antiviral response and an increase in the level of MX2. The C/C genotype with an increased frequency representation of 96.38% may be associated with more effective protection against HIV, since in this case a stronger antiviral response is activated, including MX2 stimulation. In contrast, for гомозиготыthe T/T homozygote, which has a lower frequency of 3.61%, it is possible to reduce the efficiency of antigen presentation and, as a result, a weaker response, which leads to a decrease in MX2 activity.IFNM1 is activated in response to interferons that can be induced by polymorphisms in HLA-DRA. Table 5 presents data on the expression of the IFNM1 gene in HIV1-infected patients, depending on the genotype of the rs8084 HLA-DRA polymorphism, and also includes results for the control group and for the GUSB genetic marker. The results of IFNM 1 gene1expression showed уthat in individuals with the C/C genotype of the rs8084 polymorphism of the HLA-DRA gene, in the ontrol groupe, the expression IFNM of the IFNM 1 gene was 489 ± 11.5, in HIV1 infected individuals it was 1391±18.2, GUSB 1841±8.7, in thelevel of expression relative to the GUSB gene GUSB showed an increase in expression IFNM of the IFNM 1 gene 2.8585 times in the HIV1 group of the infected population compared to the control group. Individuals with с генотип the C/T genotype of the rs 8084 polymorphism of the HLA-DRA gene, in HIV1 infected patients, the expression IFNM of the IFNM 1 gene was 1380 ± 11.1 compared to the controlой groupой 412±12.5, in relation to the control GUSB gene: 1841±8.7, the level of expression showed a significant increase in HIV1 infected patients with The C/T genotype increased by 3.3-4 times. T/T polymorphism rs 8084 of the HLA-DRA gene in the examined population in the control group was not detected, in HIV1 infected patients the values were 810±6.2, compared with the control gene GUSB 1841±8.7, the expression level had lower values compared to the C/C genotype and the heterozygous C/T genotype.Table 5. Expression parameters of the IFNM 1 gene inHIV1 infected patients depending on the genotype of the rs8084 HLA-DRA polymorphism
 |
| |
|
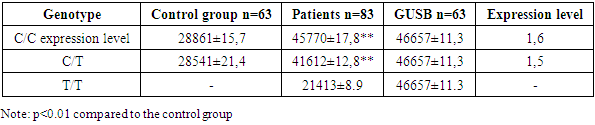
The highest level of expression of the IFNM1 gene is observed in patients with the C/T genotype 3,3,44, slightly lower in the C/C genotype 2,8989. In individuals with the T/T genotype, the expression level of the IFNM1 gene is reduced, which may indicate the influence of this genotype on gene уexpression in HIV-infectedindividuals.Thus, data analysis suggests that the level of IFNM1 gene expression in the control group also varies depending on the genotype, but in general, the level of expression in patients is significantly higher, which confirms the activation of immune responses in response to HIV-1 infection. The C/C genotype is probably associated with more active interferon stimulation and, consequently, higher levels of IFNM1. The T / T genotype may indicate a reduced response, which may also affect IFNM1 expression, reducing the effectiveness of anti-virus protection. ADAR1 is involved in RNA editing and plays an important role in immunemodulation.Table 6 provides data on the expression of the ADAR1 gene in HIV1-infected patients, depending on the genotype of the rs8084 HLA-DRA polymorphism, and also includes results for the control group and for the GUSB genetic marker.Table 6. ADAR 1 gene expression indicesin HIV1 infected patients depending on the genotype of the rs8084 HLA-DRA polymorphism
 |
| |
|
In individuals with the C/C genotype of the rs 8084 polymorphism of the HLA-DRA, gene, the expression of the ADAR1 gene in кcontrolой groupse was 28861±15.7, compared with HIV1 infected individuals 45770±17.8, GUSB 46657±11.3, in HIV1 infected individuals, the expression level of the ADAR1 gene was 1.66 times higher than in the control group groups. In HIV1 infected individuals with heterozygousom genotypeом C/T polymorphism rs 8084 of the HLA-DRA gene, ADAR1 gene expression had a 1.5-fold high level of 41612±12.8 expression compared to the control group 28541 ± 21.4, GUSB 46657 ± 11.3. The T/T genotype was not detected in the control group полиморфизма of the rs 8084 polymorphism of the HLA-DRA gene, in individuals infected with HIV1 infection, this indicator was 21413±8.9, GUSB: 46657±11.3, the expression of the ADAR1 gene with the T/T genotype in HIV1 patients had low levels compared to the C/C genotypes, and a heterozygous C/T genotype. Patients with the G/G genotype have the highest level of ADAR1 gene expression, 1.6, slightly lower in the C/T genotype, 1.5. У In individuals with the A/A genotype, the level of ADAR1 gene expression is significantly reduced, which may indicate that this genotype is associated with a lower level of this gene expression in HIV1 infected individuals.In the control group, the expression level of the ADAR1 gene is in the range of 28541±21.4 and 28861±15.7, which is the baseline value for the control group.Thus, the data in Table 6 show that the C/C and C/T genotypes are associated with an increased level of ADAR1 gene expression in patients with HIV1, while the T/T genotype is characterized by a significantly lower level of expression, which may reflect a genetic predisposition to a change in gene activity in conditions of HIV1 infection.The HLA-DRA polymorphism, which regulates the immune system response, may affect ADAR1 expression. In the case of C/C, where there is a stronger activation of the immune response, ADAR1 can be activated to a higher degree, which contributes to more effective control of viral RNA and reduced HIV replication. For T/T and C/T genotypes, where the immune response may be less pronounced, ADAR1 expression may be reduced, which leads to less effective control of viral activity. | Figure 3. Expression of the om gene MX2, IFNM1, and ADAR1 in HIV1 infected patients depending on the rs8084 HLA-DRA polymorphism |
The HLA-DRA rs8084 polymorphism affects the expression of the MX2, IFNM1, and ADAR1 genes, which play a key role in the body's antiviral defense (Figure 1). In particular, the C/C genotype is associated with a stronger immune response, increased expression of MX2, IFNM1 and ADAR1, which may contribute to a more effective fight against HIV.In contrast, the T/T genotype may be associated with a weakened immune response and reduced activity of these genes, which may contribute to a higher viral load and a more severe course of HIV infection. These studies highlight the importance of the HLA-DRA polymorphism as a possible marker of HIV predisposition and its role in the body's immune defense. These results can serve as a basis for further research in the development of personalized methods of HIV prevention and treatment based on genetic markers.
References
| [1] | Didonna A., Damotte V., Shams H., Matsunaga A., Caillier S.J., Dandekar R., Misra M.K., Mofrad M.R.K., Oksenberg J.R., Hollenbach J.A. A splice acceptor variant in HLA-DRA affects the conformation and cellular localization of the class II DR alpha-chain // Immunology. – 2021. – Vol. 162, № 2. – P. 194–207. DOI: 10.1111/imm.13273. Epub 2020 Oct 19. |
| [2] | Bosinger S.E., Utay N.S. Type I interferon: understanding its role in HIV pathogenesis and therapy // Curr HIV/AIDS Rep. – 2015. – Vol. 12, № 1. – P. 41–53. DOI: 10.1007/s11904-014-0244-6. |
| [3] | Burdick R.C., Morse M., Rouzina I., Williams M.C., Hu W.S., Pathak V.K. HIV-1 uncoating requires long double-stranded reverse transcription products // Sci Adv. – 2024. – Vol. 10, № 17. – eadn7033. DOI: 10.1126/sciadv.adn7033. Epub 2024 Apr 24. |
| [4] | Goujon C., Moncorge O., Bauby H., Doyle T., Ward C.C., Schaller T., Hue S., Barclay W.S., Schulz R., Malim M.H. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection // Nature. – 2013. – Vol. 502. – P. 559–562. |
| [5] | Goujon C., Malim M.H., et al. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells // J. Virol. – 2010. – Vol. 84. – P. 9254–9266. DOI: 10.1128/JVI.00854-10. |
| [6] | Kralovicova J., Marsh S.G., Waller M.J., Hammarstrom L., Vorechovsky I. The HLA-DRA*0102 allele: correct nucleotide sequence and associated HLA haplotypes // Tissue Antigens. – 2002. – Vol. 60, № 3. – P. 266–267. DOI: 10.1034/j.1399-0039.2002.600310.x. |
| [7] | Biswas N., Wang T., Ding M., Tumne A., Chen Y., Wang Q., Gupta P. ADAR1 is a novel multi-targeted anti-HIV-1 cellular protein // Virology. – 2012. – Vol. 422. – P. 265–277. PMID: 1965187415. |
| [8] | Peterlin B.M. Transcriptional regulation of HLA-DRA gene // Res Immunol. – 1991. – Vol. 142, № 5-6. – P. 393–399. DOI: 10.1016/0923-2494(91)90037-j. |
| [9] | Utay N.S., Douek D.C. Interferons and HIV infection: the good, the bad, and the ugly // Pathog Immun. – 2016. – Vol. 1, № 1. – P. 107–116. DOI: 10.20411/pai.v1i1.125. |





 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML




