Fatima Bakhtiyarovna Akhmedova1, Dilfuza Saburovna Matkarimova2, Kodirzhon Tukhtaboevich Boboev1
1Department of Molecular Medicine and Cellular Technologies, Republican Specialized Scientific-Practical Medical Center of Hematology MoH RUz, Tashkent, Uzbekistan
2Department of Hematology, Transfusiology and Laboratory Science, Tashkent Medical Academy, Tashkent, Uzbekistan
Correspondence to: Dilfuza Saburovna Matkarimova, Department of Hematology, Transfusiology and Laboratory Science, Tashkent Medical Academy, Tashkent, Uzbekistan.
| Email: |  |
Copyright © 2024 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Abstract
To study the contribution of IL-17F (rs763780) and IL23R (rs11209026) gene polymorphisms to the development of acute leukemia. The material for clinical and laboratory studies in the work were patients with AL (n=102) who sought diagnostic help and subsequent inpatient examination at the Republican specialized scientific - practical medical center of Hematology (RSSPMCH, Tashkent) from 2019 to 2023. Patients with AL were aged from 18 to 74 years, while the median age was 37.0±1.5 years. The diagnosis was made taking into account clinical and laboratory data. A four milliliter peripheral blood sample was taken from all participants to study single nucleotide polymorphisms IL-23R (rs11209026; G1142A) and IL-17F (rs763780; A7488G) using polymerase chain reaction of restriction fragment length polymorphism (PCR-RFLP). The samples were stored at a temperature of -80°C until DNA was isolated. The results were statistically processed using the PC application package “OpenEpi 2009, Version 2.3”. Comparing the results of the distribution of polymorphic loci of the IL17F (rs763780) interleukin gene between groups of patients with AL and healthy ones, there was a tendency to increase the risk of AL by 2.0 times and ALL by 2.0 times among carriers of the weakened Arg allele. No significant association was found between an increased risk for the formation of AL (ALL and AML) and IL23R (rs11209026) (OR<1; p>0.05).
Keywords:
IL-17F (rs763780), IL23R (rs11209026), Polymorphism, Acute leukemia
Cite this paper: Fatima Bakhtiyarovna Akhmedova, Dilfuza Saburovna Matkarimova, Kodirzhon Tukhtaboevich Boboev, Polymorphism of IL-17F (rs763780) and IL-23R (rs11209026) Genes: Role in the Development of Acute Leukemia, American Journal of Medicine and Medical Sciences, Vol. 14 No. 12, 2024, pp. 3347-3351. doi: 10.5923/j.ajmms.20241412.57.
1. Introduction
Single nucleotide polymorphisms (SNPs) are the most common type of genetic diversity in the human genome, usually associated with a harmful phenotype [11]. They can be found in different parts of genes, including promoters, exons, introns, as well as 5'- and 3'-UTRs [4]. This can lead to the formation of an altered protein, which can cause functional disruption of the protein or affect its expression, especially if they are located in coding regions or regulatory regions of genes [17].IL-23 plays an important role in the differentiation of Th17 cells and the development of pathological Th17 cells. IL-17 cytokines produced by pathological Th17 cells can induce the production of many proinflammatory cytokines and chemokines [14]. IL-17A, IL-17F and the IL-23R receptor on the cell surface in the IL-17 family not only function in chronic inflammatory diseases and autoimmune diseases [3,5,15], but also participate in tumor immunity [9]. More and more studies show that IL-17A -197G/A and IL-17F 7488T/C, two commonly studied SNPs located at the -197 IL17A promoter position and the 161 IL17F codon position, alter the function of the IL-17 protein and subsequently affect cancer susceptibility [18].Currently, many studies report the association of certain SNPs in the IL-17 gene with oncological diseases, including stomach cancer [6,8], colorectal cancer [10,12] and thyroid cancer [2]. Over the past few years, many studies have been published examining the relationship of two single-nucleotide polymorphisms with AML susceptibility [1,16,19]. One study claimed that mutant IL-17F, IL-17A genotypes did not show a significant association with the incidence of AML [7], while another showed a statistically significant increase in the homomutant IL-17F GG genotype among patients with AML compared with the control group [13]. However, the results of the study of the association of AML with polymorphism of the IL-17A, IL-17F and IL-23R genes in Chinese showed that polymorphism of the IL-17F gene was associated with susceptibility to AML [20].
2. Main Body
2.1. The Purpose of Our Research
To study the contribution of IL-17F (rs763780) and IL23R (rs11209026) gene polymorphisms to the development of acute leukemia.
2.2. Material and Methods of Study
The material for clinical and laboratory studies in the work were patients with AL (n=102) who sought diagnostic help and subsequent inpatient examination at the Republican Specialized Scientific and Practical Medical Center of Hematology (RSSPMCH, Tashkent) from 2019 to 2023. Patients with AL were aged from 18 to 74 years, while the median age was 37.0±1.5 years. The diagnosis was made taking into account clinical and laboratory data. Depending on the form of AL, the 1st combined group of patients was divided into two subgroups: 1a (n=70) - patients with acute lymphoblastic leukemia (ALL) and 1b (n=32) - patients with acute myeloblastic leukemia (AML).To carry out molecular genetic analyses of immune modulator genes, venous blood samples of 4.0 ml were taken from patients with AL and healthy into test tubes containing EDTA IL17F (rs763780) and IL23R (rs11209026), which were stored in freezers at a temperature of -80 °C until the biomaterial was completely collected.After blood collection, DNA of patients and healthy people was isolated using AmpliPrime RIBO-prep reagents (Russia) on a NanoDrop 2000 spectrophotometer (NanoDropTechnologies, USA) with a wavelength equal to A260/280 nm with a determination of DNA concentration by a ratio of 1.7/1.8.The detection of polymorphic genes IL17F (rs763780) and IL23R (rs11209026) was performed by PCR in RT mode using a RotorGene 6000/Q thermal cycler (Corbett Research).The amplification procedure was carried out by pre-denaturation of DNA at a temperature of 93°C for 1 minute; 35 denaturation cycles at a temperature of 93°C for 10 seconds; annealing of primers at a temperature of 64°C for 10 seconds; elongation process at a temperature of 72°C for 20 seconds; DNA synthesis at a temperature of 72°C for 1 minute; Separation of amplification products is carried out in a 3% agarose gel prepared on a TAE buffer by horizontal electrophoresis. To visualize the results of electrophoresis, a 1% solution of ethidium bromide is applied as a dye at the rate of 5 µl per 50 ml of molten gel. Fragments of the analyzed DNA appear as glowing orange-red stripes under UV radiation with a wavelength of 310 nm.LITEX test systems (Russia) were used to detect the studied genetic markers. The SNPs were tested for deviations from the Hardy-Weinberg equilibrium using the Chi-square test. The relative risk associated with alleles and genotypes was calculated as the odds ratio (OR) with a 95% confidence interval (CI), which were performed using the PC program “OpenEpi 2009, Version 2.3”.
2.3. Results of the Study
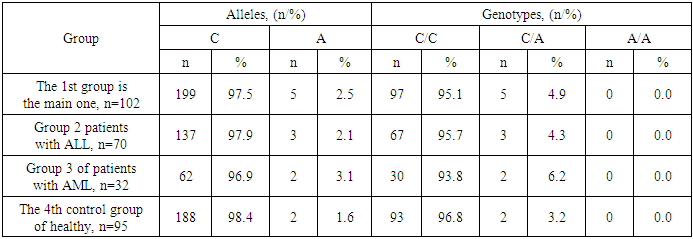
In the structure of the polymorphic marker of the IL23R (rs11209026) gene in the healthy group (n=95), along with two allelic variants G (98.4%) and A (1.6%), cases of carriage of only two genotypic variants G/G (96.8%) and G/A (3.2%) were found. Dynamics close to the structure of the IL23R (rs11209026) gene was also observed in the main group of patients with AL (n=102), in which G (97.5%) and A (2.5%), cases with only two genotypic variants G/G (95.1%) and G/A (4.9%).Along with the close frequencies of polymorphic loci of the IL23R (rs11209026) gene, carriage of a weakened homozygous A/A variant (0.0% and 0.0%) was not detected in both studied groups.Moreover, practically no carriers of the mutant Arg/Arg genotype were found in this group (see Table 1).Table 1. The frequency of polymorphic loci of the gene IL23R (rs11209026) in healthy and patients with acute leukemia
 |
| |
|
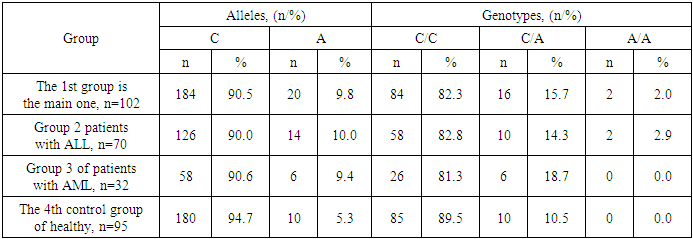
Among patients with ALL (n=70) in the structure of the IL23R (rs11209026) gene, G and A alleles were determined with frequencies of 97.9% and 2.1%, and genotypic variants G/G and G/A with frequencies of 95.7% and 4.3%, respectively.In the group of patients with AML (n=32), in the structure of the polymorphic marker of the IL23R (rs11209026) gene, the frequencies of G and A alleles were 96.9% and 3.1%, and the proportions of G/G and G/A genotypes were 93.8% and 6.2%.Accordingly, in the groups of patients with ALL and AML, the carriage of a weakened homozygous variant A/A was also not established (0.0% and 0.0%).Thus, evaluating the structural features of the gene IL17F (rs763780) in the groups of patients and healthy, an undefined increase in the frequencies of weakened allele A and genotype G/A among patients was found, despite the fact that their frequency was most noticeably higher among patients with AML.To clarify the possible relationship between IL23R (rs11209026) gene polymorphism and the risk of AL, we compared the degree of differences between the established frequencies of allelic and genotypic variants in the patient and healthy groups. Comparing the differences in the main group with AL compared with healthy ones, it was found that the weakened allele A (2.5% vs. 1.1%; χ2=1.1; P=0.3; OR=2.4; 95%CI: 0.47-11.7) and genotype G/A (4.9% vs. 2.1%; χ2=1.1; P=0.3; OR=2.4; 95%CI: 0.48-12.1) they were 2.4 times more common among patients with AL, but the differences between the groups in their distribution did not reach statistically significant values.Thus, compared with the control group, in AL, the carrier has weakened variants of the A allele (χ2=1.1; P=0.3) and the genotype G/A (χ2=1.1; P=0.3) for the IL23R (rs11209026) gene did not differ significantly, which serves as evidence that the studied genetic marker alone cannot be associated with the risk of AL.Comparing the differences in the distribution of loci according to the polymorphism of the IL23R (rs11209026) gene in the group of patients with ALL compared with healthy, an unreliable increase in the frequencies of attenuated A allele was found (2.1% vs. 1.1%; χ2=0.6; P=0.5; OR=2.1; 95%CI: 0.35-12.0) and genotype G/A (4.3% vs. 2.1%; χ2=0.7; P=0.5; OR=2.1; 95%CI: 0.35-12.3) 2.1 times at ALL.Similar statistically insignificant differences in the distribution of loci according to the polymorphism of the IL23R (rs11209026) gene compared with healthy ones were also revealed in the group of patients with AML, in which the frequencies of the weakened A allele (3.1% vs. 1.1%; χ2=1.3; P=0.3; OR=3.0; 95%CI: 0.46-20.0) and genotype G/A (6.3% vs. 2.1%; χ2=1.3; P=0.3; OR=3.1; 95%CI: 0.46-20.9) they were more frequent by 3.0 and 3.1 times, respectively.Thus, compared with the control group for the IL23R (rs11209026) gene, the groups of patients with ALL and AML carried weakened variants of the A allele (χ2=0.6; P=0.5 and χ2=1.3; P=0.3) and genotype G/A (χ2=0.7; P=0.5 and χ2=1.3; P=0.3) did not differ significantly. In turn, this proved that the studied genetic marker alone cannot be associated with the risk of ALL and AML.In the frequencies of polymorphic loci of the IL23R (rs11209026) gene between the groups of patients with ALL and AML, the differences were also not statistically significant for the weakened A allele (2.1% vs. 3.1%; χ2=0.2; P=0.7; OR=0.7; 95%CI: 0.11-4.12) and genotype G/A (4.3% vs. 6.3%; χ2=0.2; P=0.7; OR=0.7; 95%CI: 0.11-4.19) when their frequencies exceed among patients with AML by less than one.Thus, comparing the results of the distribution of polymorphic loci of the IL23R (rs11209026) gene between the groups of patients and healthy, no statistically significant differences were found. Compared with healthy patients, an unreliable increase in the frequencies of weakened allele A and G/A loci was found in the main group by 2.4 times (χ2=1.1; P=0.3 and χ2=1.1; P=0.3, respectively), in the group of patients with ALL by 2.1 times (χ2=0.6; P=0.5 and χ2=0.7; P=0.5, respectively) and with AML at 3.0 (χ2=1.3; P=0.3) and 3.1 times (χ2=1.3; P=0.3) accordingly.Further analyzing the structural features of the polymorphic marker of the IL17F (rs763780) gene in the healthy group (n=95), the frequencies of allelic (His = 94.7% and Arg = 5.3%) and genotypic variants (His/His = 89.5%, His/Arg= 10.5%, Arg/Arg = 0.0%) were determined among which, the proportion of the main loci was determined more often than the weakened variants. Moreover, practically no carriers of the mutant Arg/Arg genotype were found in this group (see Table 2).Table 2. The frequency of polymorphic loci of the gene IL 17F (rs763780) in healthy and patients with acute leukemia
 |
| |
|
The structure of the polymorphic marker of the IL17F (rs763780) gene in the main group of patients (n=102) was characterized by lower frequencies of the main loci (His = 90.2% and His/His = 82.3%) and higher frequencies of attenuated variants (Arg = 9.8% and His/Arg= 15.7%). Moreover, in the main group, Arg/Arg mutant genotype was detected in 2% of cases.In the group of patients with ALL (N=70), the structure of the polymorphic marker of the IL17F (rs763780) gene was characterized by the lowest frequency of its main allele (90.0%) and the highest proportion of a weakened variant of Arg (10.0%) among all studied categories. I believe that I am in a group with a very large population with three possible answer options (His/His = 82.8%, His/Arg= 14.3%, Arg/Arg = 2.9%).At the same time, in the group of patients with AML (n=32), in the structure of the polymorphic marker of the IL17F (rs763780) gene, along with a relatively lower frequency of the main allele of His (90.6%) and a weakened variant of Arg (9.4%), compared with similar ones in the group with ALL, a lower frequency of carrier of the main genotype (His/His = 81.3%) with the highest frequency of the heterozygous variant (His/Arg=18.7%). At the same time, no cases with the mutant Arg/Arg genotype were detected in this group (0.0%).Thus, the structure of the IL 17F (rs763780) gene in the studied groups was characterized by a higher frequency of carriage by unfavorable loci among patients and the presence of carriage of mutant homozygotes only among the group of patients with ALL. At the same time, the established high frequency of weakened loci in patient groups compared with the healthy group may be associated with their participation in an increased risk of AL.To determine the strength of correlations between the studied polymorphism loci of the IL17F (rs763780) gene and the risk of AL, a comparative assessment of the degree of differences between the established frequencies of allelic and genotypic variants in the main group of AL compared with those in the healthy group was carried out. Thus, between the frequencies of the mutant Arg allele, a tendency was found to increase its proportion among the main group with AL by 2.0 times (9.8% vs. 5.3%; χ2=2.9; P=0.1; OR=2.0; 95% CI: 0.9 - 4.25). Along with this, between the frequencies of His/His genotypes (82.4% vs. 89.5%; χ2=2.0; P=0.2; OR=0.5; 95%CI: 0.24-1.25) and His/Arg (15.7% vs. 10.5%; χ2=1.1; P=0.3; OR=1.6; 95%CI: 0.68-3.66) there were no significant differences in the studied groups.Thus, compared with the control group, among carriers of the mutant attenuated Arg allele, there is a tendency to increase the risk of AL by 2.0 times (χ2=2.9; P=0.1). Therefore, the Arg allele of the IL 17F (rs763780) gene can be considered a genetic marker that increases the risk of AL.Assessing the strength of correlations between the studied polymorphism loci of the IL17F (rs763780) gene and the risk of ALL compared with the control, a weak tendency to increase the frequency of the mutant Agd allele in ALL by 2.0 times was found (10.0% vs. 5.3%; χ2=2.7; P=0.2; OR=2.0; 95%CI: 0.87 - 4.58). However, as in the main group, the difference between the frequencies of His/His genotypes (82.9% vs. 89.5%; χ2=1.5; P=0.3; OR=0.6; 95%CI: 0.23-1.39) and His/Arg (14.3% vs. 10.5%; χ2=0.5; P=0.5; OR=1.4; 95%CI: 0.56-3.6) no significant differences were found in the studied groups.Consequently, the detected higher frequency of the unfavorable Arg allele among the group of patients with ALL compared with the control shows a tendency to increase the risk of ALL by 2.0 times (χ2=2.7; P=0.2).At the same time, despite the large proportion of weakened IL17F (rs763780) gene polymorphism loci in patients with AML compared with controls, there was no statistically significant association between them and the risk of AML. For example, among patients with AML, although the frequencies of the weakened Arg allele and His/Arg genotype were 1.9 times higher than the control frequencies (9.4% vs. 5.3%; χ2=1.4; P=0.3; OR=1.9; 95%CI: 0.66 - 5.27) and 2.0 times (18.8% vs. 10.5%; χ2=1.5; P=0.3; OR=2.0; 95%CI: 0.66 - 5.83) there were no statistically significant differences between the groups in their proportion. A similar absence of statistically significant differences between the studied groups was observed in the frequencies of the main His allele (90.6% vs. 94.7%; χ2=1.4; P=0.3; OR=0.5; 95%CI: 0.19-1.52) and His/His genotype (81.3% vs. 89.5%; χ2=1.5; P=0.3; OR=0.5; 95%CI: 0.17-1.51).Consequently, the detected higher frequency of unfavorable Arg loci (χ2=1.4; P=0.3) and His/Arg (χ2=1.5; P=0.3) is not associated with the risk of AML.
3. Conclusions
Comparing the results of the distribution of polymorphic loci of the IL17F (rs763780) interleukin gene between groups of patients and healthy individuals, there was a tendency to increase the risk of AL by 2.0 times (χ2=2.9; P=0.1) and ALL by 2.0 times (χ2=2.7; P=0.2) among carriers of the weakened Arg allele. Comparing the results of the distribution of polymorphic loci of the IL23R (rs11209026) gene between the groups of patients and healthy, no statistically significant differences were found.
References
| [1] | Alnagar A. A. et al. Prognostic Value of Interleukin-17 A Gene Polymorphism and Serum IL-17 Levels in Adult Acute Myeloid Leukaemia Patients // The Egyptian Journal of Hospital Medicine. – 2020. – Т. 81. – №. 2. – С. 1352-1358. |
| [2] | Bertol B. C. et al. Polymorphisms at the IL17A and IL17RA genes are associated with prognosis of papillary thyroid carcinoma // Archives of Medical Research. – 2022. – Т. 53. – №. 2. – С. 163-169. |
| [3] | Botros S. K. A., Ibrahim O. M., Gad A. A. Study of the role of IL-17F gene polymorphism in the development of immune thrombocytopenia among the Egyptian children // Egyptian Journal of Medical Human Genetics. – 2018. – Т. 19. – №. 4. – С. 385-389. |
| [4] | Deng N. A. et al. Single nucleotide polymorphisms and cancer susceptibility // Oncotarget. – 2017. – Т. 8. – №. 66. – С. 110635. |
| [5] | Elfasakhany F. M., Eldamarawi M. A., Khalil A. E. Association between interleukin-17 gene polymorphism and rheumatoid arthritis among Egyptians // Meta Gene. – 2018. – Т. 16. – С. 226-229. |
| [6] | Elshazli R. M. et al. Genetic polymorphisms of IL-17A rs2275913, rs3748067 and IL-17F rs763780 in gastric cancer risk: evidence from 8124 cases and 9873 controls //Molecular biology reports. – 2018. – Т. 45. – С. 1421-1444. |
| [7] | Elsissy M. et al. Interleukin-17 gene polymorphism is protective against the susceptibility to adult acute myeloid leukaemia in Egypt: a case-control study // Open Access Macedonian Journal of Medical Sciences. – 2019. – Т. 7. – №. 9. – С. 1425. |
| [8] | Gao J. F. et al. Associations of the IL-17A rs2275913 and IL-17F rs763780 polymorphisms with the risk of digestive system neoplasms: A meta-analysis // International Immunopharmacology. – 2019. – Т. 67. – С. 248-259. |
| [9] | Hu Y. et al. Associations of IL-17A-197G/A and IL-17F 7488T/C polymorphisms with cancer risk in Asians: An updated meta-analysis from 43 studies // Gene. – 2021. – Т. 804. – С. 145901. |
| [10] | Javadirad E. et al. Associations of IL-4, IL-4R, IL-17A, and IL-17F polymorphisms with colorectal cancer risk: a meta-analysis, meta-regression, and trial sequential analysis // Journal of Interferon & Cytokine Research. – 2022. – Т. 42. – №. 5. – С. 203-219. |
| [11] | Lee M. H. et al. Interleukin 17 and peripheral IL-17-expressing T cells are negatively correlated with the overall survival of head and neck cancer patients // Oncotarget. – 2018. – Т. 9. – №. 11. – С. 9825. |
| [12] | Li G. et al. The associations between interleukin-17 single-nucleotide polymorphism and colorectal cancer susceptibility: a systematic review and meta-analysis // World Journal of Surgical Oncology. – 2022. – Т. 20. – №. 1. – С. 116. |
| [13] | Mahfouz K., Abo-Nar A., Bendary S. Role of Interleukin17F (IL17F) Gene Polymorphism in Susceptibility to Acute Myeloid Leukemia // J Leuk. – 2018. – Т. 7. – С. 252-56. |
| [14] | Mills K. H. G. IL-17 and IL-17-producing cells in protection versus pathology // Nature Reviews Immunology. – 2023. – Т. 23. – №. 1. – С. 38-54. |
| [15] | Mohammed F. N. et al. Study of interleukin-17 gene polymorphism and susceptibility to vitiligo in a sample of the Egyptian population // Journal of the Egyptian Women’s Dermatologic Society. – 2017. – Т. 14. – №. 1. – С. 45-48. |
| [16] | Nasr A. S. et al. IL 17F and IL-23 gene polymorphism in patients with acute myeloid leukemia, an Egyptian study // Journal of Bioscience and Applied Research. – 2018. – Т. 4. – №. 4. – С. 469-477. |
| [17] | Ogawa H. et al. Somatic mosaicism in biology and disease // Annual review of physiology. – 2022. – Т. 84. – №. 1. – С. 113-133. |
| [18] | Samiei G. et al. Association between polymorphisms of interleukin-17A G197A and interleukin-17F A7488G and risk of colorectal cancer // Journal of Cancer Research and Therapeutics. – 2018. – Т. 14. – №. Suppl 2. – С. S299-S305. |
| [19] | Zayed R. A. et al. IL-17 a and IL-17 F single nucleotide polymorphisms and acute myeloid leukemia susceptibility and response to induction therapy in Egypt // Meta Gene. – 2020. – Т. 26. – С. 100773. |
| [20] | Zhu B. et al. Correlation between acute myeloid leukemia and IL-17A, IL-17F, and IL-23R gene polymorphism [Retraction] // Int J Clin Exp Pathol. – 2020. – Т. 13. – №. 7. – С. 1932. |


 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML
