-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2024; 14(12): 3149-3153
doi:10.5923/j.ajmms.20241412.13
Received: Nov. 21, 2024; Accepted: Dec. 9, 2024; Published: Dec. 10, 2024

Results of an Analysis of the Prevalence of the rs20417 Polymorphism in the COX2 Gene Among Patients with Chronic Polyposis Rhinosinusitis and Chronic Rhinosinusitis and in the Control Group
Ulugbek S. Khasanov, Ilyosjon O. Soatov, Shoximardon X. Xodjanov, Nazim O. Axundjanov, Gulnora A. Raximjanova
Tashkent Medical Academy, Tashkent, Uzbekistan
Correspondence to: Ilyosjon O. Soatov, Tashkent Medical Academy, Tashkent, Uzbekistan.
| Email: |  |
Copyright © 2024 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Differences between SRSwP, SRS and control group patients were noted at the trend level. Analysis of the contribution of the COX2 gene polymorphism to the frequency of occurrence, development and clinical course. The study was conducted using the method of molecular genetic testing, in which 116 people aged 18-60 years from chronic polyposis rhinosinusitis, chronic rhinosinusitis and control group participated. The prevalence of the rs20417 76 G/C polymorphism in the COX2 gene was analyzed among patients with chronic polyposis rhinosinusitis and chronic rhinosinusitis and in the control group. We can conclude that there are no significant differences in the frequency of detection of allelic and genotypic variants of the 76 G/C polymorphic locus in the COX2 gene.
Keywords: Chronic polyposis rhinosinusitis, Chronic rhinosinusitis, Polymorphism, Encoded gene, Inflammatory response, COX2 gene, Homozygous
Cite this paper: Ulugbek S. Khasanov, Ilyosjon O. Soatov, Shoximardon X. Xodjanov, Nazim O. Axundjanov, Gulnora A. Raximjanova, Results of an Analysis of the Prevalence of the rs20417 Polymorphism in the COX2 Gene Among Patients with Chronic Polyposis Rhinosinusitis and Chronic Rhinosinusitis and in the Control Group, American Journal of Medicine and Medical Sciences, Vol. 14 No. 12, 2024, pp. 3149-3153. doi: 10.5923/j.ajmms.20241412.13.
1. Introduction
- SRSwP is a complex multifactorial disease that involves several genetic, immunological, environmental, and mucosal changes in the body, but still the etiology remains unclear. Many potential factors have been identified, such as various allergic reactions, impaired secretion of mucous membranes, decreased immunity, impaired epithelial protection, exposure to microbes and the environment. Further studies are needed to determine the role of genetic factors and their interaction with these factors in the pathophysiology of SRSwP disease. The lack of appropriate animal models, difficulties in standardizing phenotypes, the need for large controlled cohorts, and the high cost and low reproducibility of studies are major obstacles to elucidating the pathophysiology of SRSwP [5,16].There is convincing evidence of the influence of genetic factors on the pathophysiology of SRSwP. Mutations in the cystic fibrosis transmembrane regulator (CFTR) gene cause cystic fibrosis (CF), which is the most frequently duplicated gene associated with SRS. There is a high prevalence of SRSwP in cystic fibrosis carriers, but some studies show that CFTR mutations also occur in SRSwP patients without cystic fibrosis [14,8]. Family studies suggest a genetic factor in the pathogenesis of SRSwP, but environmental factors also play an important role in the development of nasal polyps. For example, observations have shown that even twins developed from the same egg have been studied to have different polyp tissue [18]. This part of the work is devoted to the study of the distribution frequencies of polymorphism of the COX2 gene, as well as to the analysis of the contribution of this polymorphism to the occurrence, development and clinical course of SRSwP and SRS [5,23].
2. Materials and Methods
- In 2023-2024, clinical studies were conducted on 85 patients with chronic rhinosinusitis (with polyps and without polyps), who were examined and treated in the ENT departments of the multidisciplinary clinic of the Tashkent Medical Academy and the private clinic "Tashkent LOR Center" in accordance with the purpose of the research and to fulfill the set tasks. The control group included 31 healthy volunteers aged from 18 to 60 among the employees of the private clinic "Tashkent LOR Center". All volunteers included in the study did not have acute diseases, especially infectious diseases and chronic inflammatory pathologies during the month before the start of the study.DNA code information is given to us for life, so we have a growing interest in genetic markers. Recurrence in another person in the family of a patient with SRSwP due to genetic predisposition is 15-50%. When genetic predisposition was studied by American researchers, it was found that; the first-degree relative of an infected patient was 4.1 times more likely to be infected, and the second-degree relatives were 3.3 times more likely to be infected. In the research of Swedish scientists, it was found that the frequency of recurrence in the family of an infected patient is 5 times higher than in the rest of the population. In another study by scientists, monozygotic twins were observed, in this case, SPRS recurrence was not observed, from which we can conclude that the role of external environmental factors is not always dependent on heredity [10,27].In polyp tissues and intranasal secretions, an increase in the concentration of various inflammatory mediators, in particular interleukins, is observed due to an increase in their synthesis by de novo effector cells [29]. Eosinophils (IL-4, IL-12, IL-13, GM-CSF), the main pro-inflammatory cytokines (IL-1, IL-2, FNO-α, IL-10) that contribute to the chronic inflammation in the nasal cavity, regulator special importance is given to the increase in the concentration of cytokines involved in the development, recruitment and activation of cytokines (IL-10, TLR2b) [8,3].The distribution of allele gene frequencies of resistance predisposition, which forms the risk of development or resistance to multifactorial pathology, is determined by the racial and ethnic origin of the studied group [18].Cytokine system is a universal regulatory network of mediators designed to control reproduction, differentiation and functional activity of cellular elements in all homeostatic systems of the body, depending on which the immune system plays a very important role in the occurrence and development of diseases [12].Cytokines - leukocyte mediators of inflammation are included in pro-inflammatory or anti-inflammatory regulatory peptides. IL-1 and TNF-α play the biggest role in pro-inflammatory processes. The mechanisms by which these cytokines participate in the inflammatory response are largely common [13]. They can increase the expression of the COX-2 gene with the increase in the production of leukotrienes and prostaglandins and their inclusion in the pathological process, thereby increasing the migration of leukocytes and inflammatory infiltration and activating the endothelium. IL-6 and IL-8 also have anti-inflammatory properties [2,21]. Anti-inflammatory cytokines are active antagonists of IL-1 and TNF-α, the most active of which are IL-4 and IL-10. Their anti-inflammatory activity is manifested by suppressing the production of IL-1 and TNF-α, colony-stimulating factors, and reducing the cytotoxicity of macrophages [24].
3. Result and Discussions
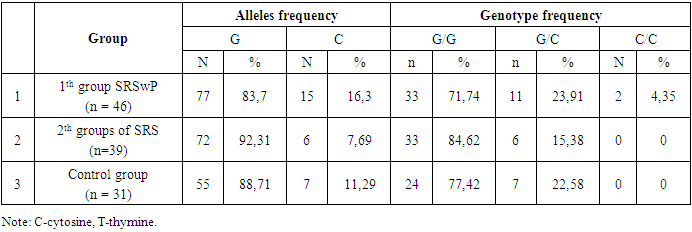
- The prevalence of the rs20417 76 G/C polymorphism in the COX2 gene was analyzed among patients with chronic polyposis rhinosinusitis and chronic rhinosinusitis and in the control group. The distribution of alleles and genotypes of the 76 G/C polymorphism in the COX2 gene among patients with SRSwP, SRS and conditionally healthy people was studied, the results of which are presented in Table 1.
|
|
|
4. Conclusions
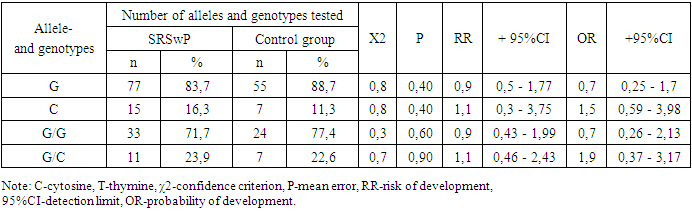
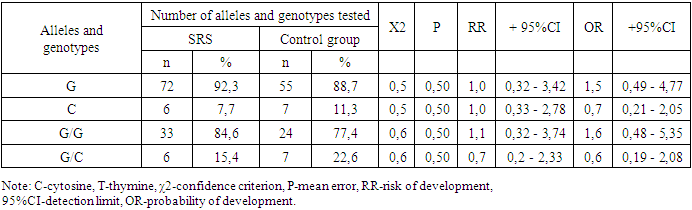
- Summarizing the obtained results, we can conclude that there are no significant differences in the frequency of detection of allelic and genotypic variants of the 76 G/C polymorphic locus of the COX2 gene in the 1st and 2nd groups of patients and the control group. There is no significant contribution of this locus in the formation and development of any form of SRS. However, there is a significant trend toward an increased frequency of the G/G genotype in patients with SRSwP compared with controls.The non-significant detection of this genotype in patients with SRSwP compared to SRS confirms the insignificant protective role of the genotypic variant of the 76 G/C polymorphism in the COX2 gene with respect to the development of SRSwP in patients (χ2=0.7; R=0.9; RR=1.1; OR=1.9 ; 95% CI: 0.46–2.43).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML