-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2024; 14(11): 2981-2984
doi:10.5923/j.ajmms.20241411.67
Received: Nov. 5, 2024; Accepted: Nov. 25, 2024; Published: Nov. 27, 2024

Metod for Rapid Isolation of Methicillin Resistant Staphylococcus Aureus Strains from Biological Materials
Murodova Ijobatxon Abdulbosi Qizi1, Ibragimov Adil Ahmedovich2
1Basic Doctoral Student, Tashkent Scientific Research Institute of Vaccines and Serums, Uzbekistan
2Doctor of Biological Sciences, Head of the Laboratory of High Technologies, Tashkent Scientific Research Institute of Vaccines and Serums, Uzbekistan
Correspondence to: Murodova Ijobatxon Abdulbosi Qizi, Basic Doctoral Student, Tashkent Scientific Research Institute of Vaccines and Serums, Uzbekistan.
| Email: |  |
Copyright © 2024 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Controlling the incidence of methicillin-resistant staphylococcus aureus (MRSA) in developed countries is a key aspect of the prevention and effective treatment of many severe infections caused by nosocomial and other purulent inflammation. In addition, the detection rate of MRSA strains also plays a role in this process. Currently, in clinical microbiology, the identification of MRSA using traditional bacteriological methods takes a long time. Recently, since bacteriological culturing of MRSA involves a 2-3-day delay until final results are obtained, rapid detection methods using PCR techniques have been developed. Performing PCR tests reduces the detection time of MRSA patients from 48-72 to 2-5 hours. Clinical evaluation data showed that MRSA can be detected with very high sensitivity in molecular genetic studies.In this study, we will try to evaluate the benefits of MRSA detection by PCR. 43 Staphylococcus aureus strains isolated by the classical method were selected for PCR analysis (based on the results of PCR, methicillin-resistant Staphylococcus aureas MRSA (N = 23) and methicillin-sensitive Staphylococcus aureas MSSA (N = 20) were selected. Diagnostic values of the analysis showed high sensitivity. This real-time PCR analysis has proven to be a fast, sensitive, and specific tool for systematic detection of MRSA.Centralized hospitals now have PCR in clinical laboratories, and using it to detect MRSA can help reduce MRSA identification time and contribute to the success of infection control programs in hospitals.
Keywords: Staphylococcus aureus, Methicillin resistance, MRSA, mecA, Molecular detection, PCR
Cite this paper: Murodova Ijobatxon Abdulbosi Qizi, Ibragimov Adil Ahmedovich, Metod for Rapid Isolation of Methicillin Resistant Staphylococcus Aureus Strains from Biological Materials, American Journal of Medicine and Medical Sciences, Vol. 14 No. 11, 2024, pp. 2981-2984. doi: 10.5923/j.ajmms.20241411.67.
1. Introduction
- Treatment of diseases caused by the methicillin-resistant bacterium Staphylococcus aureus (MRSA) is one of the most urgent medical problems worldwide. WHO list of priority bacterial pathogens for 2024 [10] divides pathogenic bacteria into important groups with high and medium priority. Methicillin-resistant Staphylococcus aureus is included in the highest group, the priority areas of practical medicine remain the diagnosis of this pathogen, the study of antibiotic resistance, and the improvement of effective treatment methods and preventive measures.Despite numerous scientific studies and the availability of domestic and foreign antibiotics for the prevention and treatment of bacterial infections worldwide, the incidence of MRSA has increased significantly in recent years. In some parts of the African and American regions, about 80% and 90% of Staphylococcus aureus injections are resistant to methicillin (MRSA), respectively, and standard antibiotic treatment does not work [7]. Staphylococcus aureus causes infections with significant mortality rates, including superficial and deep skin infections, soft tissue infections, bacteremia, myocarditis, osteomyelitis, pneumonia, toxic shock syndrome, and staphylococcal scaly skin syndrome.For the first time, gold methicillin-resistant staphylococci were discovered in England in 1961 and identified a year after the introduction of the antibiotic into clinical practice [1]. Resistance of Staphylococcus aureus to methicillin at the genetic level is associated with the presence of the SCCmec complex, the staphylococcus chromosomal cassette. This complex may include mecA or mecc (whose function is to encode the synthesis of an additional penicillin-binding protein); regulatory elements-meci and mecr 1 (control of mecA or mecc transcription); additional mec-bound DNA sequences, transposons, insertion elements, and other antibiotic resistance genes. To date, 12 different types of staphylococcal chromosomal cassettes have been described by size, location, and the presence of certain structural elements [8].Antibiotic resistance of MRSA strains is characterized by the presence of the mecA gene found in the bacterium. This gene, as a mobile genetic element and encoding a low level of penicillin-binding protein (pbp2-a), is resistant to many antibiotics, including penicillin and other beta-lactam antibiotics. The structural gene that is the determinant encoding pbp2a mecA Staphylococcus aureus in the calculated methicillin resistance is considered a molecular symbol [4]. The mecA gene can be identified by PCR, The gold standard for determining methicillin resistance is selected [2]. The results of the traditional method with bacterial culture, identification and antimicrobial sensitivity tests last from 48 to 72 hours. Rapid identification of the specific mechanism of resistance in molecular testing allows treating physicians to avoid potentially undesirable treatments. To quickly detect methicillin-resistant Staphylococcus aureus (MRSA) colonization in the throat and nose of patients, clinical microbiology laboratories should choose PCR methods or classical methods. Multiple chromogenic and differential MRSA Feed agars have been shown to produce results within 18-24 hours [9]. Results within 2-3 hours, antibiotic resistance genes provide important targets for molecular detection methods [3].The sale of antibacterial drugs in our country, weak local epidemiological control in empirical treatment demonstrate the great importance of this problem.The relevance and increasing relevance of the problems associated with this situation in diagnosis and treatment is also due to the resistance to antibacterial therapeutic drugs.The aim of the study is to identify methicillin-resistant Staphylococcus aureus (MRSA). rt-PCR and molecular-genetic assessment of its distribution dynamics.
2. Materials and Methods
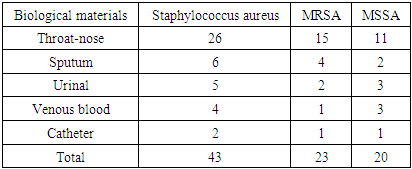
- As an object of research, in the bacteriological laboratory of the Republican Scientific Center for Emergency Medicine (RCMP) in May-June 2024, 43 Staphylococcus aureus isolates were obtained, isolated by the classical method from patients receiving inpatient treatment. As a subject of the study, the patient has 26 pieces of throat and nose smears, 6 pieces of sputum, 5 pieces of gobi, 4 pieces.venous blood, 2 pieces of catheter grease, as biomaterials are taken.).Research methods. Microbiological (microscopic, tinctorial, cultural, biochemical) and molecular-genetic (polymerase chain reaction) methods of testing sensitivity to antibiotics (discodiffusion method) were used to solve research problems and achieve the goal. Microbiological tests are performed in the bacteriological laboratory of the Republican Scientific Center of Emergency Medicine and the Tashkent Microbiological Laboratory of Vaccines and Serum, as well as detection by RT-PZR Tashkent vaccine-serum iti was performed in a high-tech laboratory.Microbiological method. All biological material is initially found in blood agar 37֯ Grown in C for 24 hours. Staphylococcus aureusfrom blood agar β-strains from colonies that have undergone hemolysis biochemical tests according to the activity of coagulase, catalase, yellow mannitol fermentation MRSA and mssa (methicillin-sensitive Staphylococcus aureus) the basis of the strains was a preservative medium tryptachinase with the addition of 3% glycerol, which collected in a nutrient medium.Test for sensitivity to antibiotics. Staphylococcus aureus colonies identified as Muller Hilton in feed media by disk diffusion in accordance with EUKACT rules were performed on the following disks: ampecillin, benzylpecillin, gatifloxacin, ofloxacin, moxifloxacin, cefazolin, cefuroxime, cefoxitin, cefaperazone, erythromycin, roxithromycin.RT-PZR method. The collected 43 isolates were tested «АмплиСенс-MRSA-скрин-титр-FL» in Russia using RT-PCR. Recording of the fluorescence signal was performed directly during PCR, using an amplifier with a “real-time” detection system. The reagent kit contains an internal control sample, which was added to each test sample during the DNA extraction step, allowing control of the analysis process for each sample. A “Hot start” is used in the set of reagents, which significantly reduces thr number of non-specific reactions. “Hot start” is provided by using a chemically modified TAQ polymerase. Chemically modified polymerase.
3. Results
- A total of 43 isolates of the Republican Scientific Center of Emergency Care were assembled from different departments, including women 48%, men 52%, mainly in the age groups from 0 to 20 years (65.11%), from 20 to 40 years (20.9%) to 41-60 years (6.9%) and over 60 years (6.9%). (Diagram 1)
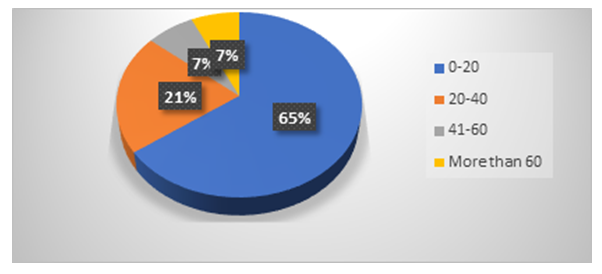 | Diagram 1. Staphylococcus aureus frequency of occurrence of in the younger groups |
|
|
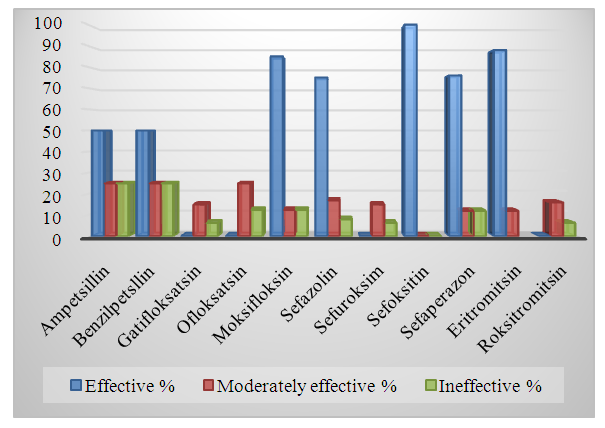 | Diagram 2. Staphylococcus aureus is an indicator of sensitivity to antibiotics |
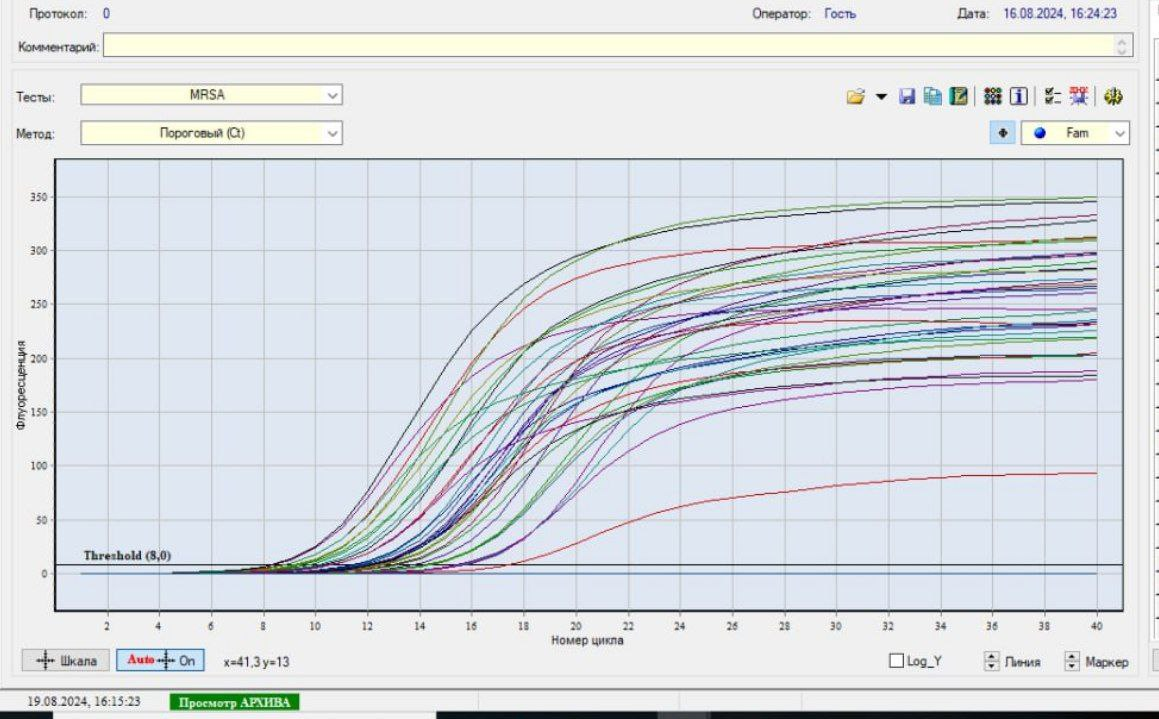 | Picture 1. RT-PZR method results |
4. Discussions
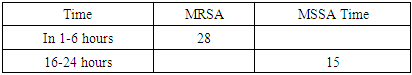
- It is known that S. aureus, unlike many other Staphylococcus species, has sufficiently well-defined derivatives and enzymatic properties, which practically does not cause difficulties in identifying it. However, the scientific literature notes that MRSA variants belonging to this type often also exhibit a typical biochemical properties among antibacterial drugs, which can be a source of diagnostic errors [5]. S.aureus is a major pathogen causing a wide spectrum of Clinical manifestations and beta-lactam antibotics are the Drugs of Therapeutic Choice. Since the introduction of methicillin into clinical use on 1961, the occurrence of MRSA strains has enormously increased and MRSA is now one of the most important nosocomial pathogenes worldwide [6]. When we analyze by age S.aureus whether S. aureus infectionsare primarily breastfed in the under-20 age group, especially in young children, complications are fatal. (Table 1, Diagram 1). In our study, we can see that MRSA strains are not only resistant to methicelin, but also distributed in heterogeneous organs, and their colonization mainly occurs in the mucous membranes of the throat and nose. (Table 2). In the classical style, we can see the resistance of MRSA strains not only to metacillin (oxacillin) antibiotics, but also to other cephalosporins (cefoxitin of the 2nd generation with a 100% result) (Table 2, Diagram 2). While bacteriological coagulase tests resolve rabbit citrate plasma clotting within 1-6 hours for MRSA strains at 24-hour intervals, (Table 2) 48-72 hours are required for culturing MRSA strains, identification, and determination of antibiotic sensitivity. During this period, the dangerous consequences of diseases caused by MRSA may increase.
5. Conclusions
- Based on the above, simultaneous detection of MRSA strains by fast and effective methods using mecA genes gives positive results. The diagnostic values of the analysis turned out to be highly sensitive and specific tools. Real-time PCR is currently available in many laboratories, so using it to detect MRSA strains can help shorten the duration of test results and control infectious diseases in hospitals.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML