-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2024; 14(9): 2322-2327
doi:10.5923/j.ajmms.20241409.42
Received: Aug. 19, 2024; Accepted: Sep. 8, 2024; Published: Sep. 21, 2024

Analysis of HLA-DRA Gene Polymorphism in Uzbek Population with HIV Infection
Shodiyeva D.1, Yarmukhamedova N. A.1, Qodirov J. F.1, Ziyadullayev Sh. Kh.2
1Samarkand State Medical University, Samarkand, Uzbekistan
2Institute of Immunology and Human Genomics of the Academy of Sciences, Tashkent, Uzbekistan
Correspondence to: Shodiyeva D., Samarkand State Medical University, Samarkand, Uzbekistan.
| Email: |  |
Copyright © 2024 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

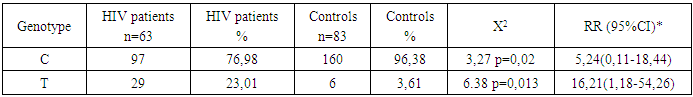
This study investigates the polymorphisms of the HLA-DRA gene, particularly focusing on the C and T alleles, in the Uzbek population with HIV infection. A cohort of 63 HIV-positive individuals and 83 healthy controls from the Samarkand region were genotyped using PCR-RFLP. The study revealed a significant association between the T allele and an increased risk of HIV infection, with the T/T genotype showing the strongest correlation. The T allele was more prevalent in the HIV-positive group (23.01%) compared to the control group (3.61%), suggesting a genetic predisposition to HIV. Conversely, the C allele was less frequent in the HIV-positive group (76.98%) compared to the control group (96.38%), indicating a potential protective effect. These findings underscore the importance of genetic factors in HIV susceptibility and suggest that HLA-DRA polymorphisms could serve as valuable markers for identifying individuals at higher risk. This study contributes to the understanding of the genetic epidemiology of HIV and highlights the potential for personalized medicine approaches in managing the disease.
Keywords: HLA-DRA, HIV, Polymorphism, Genetic susceptibility, Uzbek population
Cite this paper: Shodiyeva D., Yarmukhamedova N. A., Qodirov J. F., Ziyadullayev Sh. Kh., Analysis of HLA-DRA Gene Polymorphism in Uzbek Population with HIV Infection, American Journal of Medicine and Medical Sciences, Vol. 14 No. 9, 2024, pp. 2322-2327. doi: 10.5923/j.ajmms.20241409.42.
1. Introduction
- The Human Leukocyte Antigen (HLA) system is a critical component of the adaptive immune response, responsible for presenting foreign peptides to T-cells, thereby initiating an immune response. Among the various classes of HLA molecules, the class II molecules play a pivotal role in presenting extracellular antigens to CD4+ T-cells. The HLA-DRA gene, located on chromosome 6, encodes the alpha chain of the HLA-DR molecule, one of the most important class II molecules. This alpha chain pairs with the beta chain encoded by various DRB genes to form a heterodimer that is essential for the immune system’s ability to recognize and respond to pathogens [1,2].HLA-DRA is particularly noteworthy for its limited polymorphism, especially in the peptide-binding region, which contrasts with the high variability seen in other HLA genes such as HLA-DRB1. The limited variation in HLA-DRA suggests that it plays a fundamental and conserved role in immune function across populations. The gene is expressed predominantly on the surface of antigen-presenting cells (APCs) like B-lymphocytes, dendritic cells, and macrophages, which are critical for initiating and modulating immune responses. The primary function of these APCs is to capture, process, and present antigens to T-cells, facilitating the recognition of foreign invaders such as bacteria, viruses, and other pathogens [3,4,5].In the context of infectious diseases such as HIV, the role of the HLA system becomes even more crucial. HIV, the virus responsible for AIDS, specifically targets the immune system, leading to a progressive decline in immune function. The interaction between HLA molecules and T-cells is a key determinant of the immune response to HIV infection. Previous studies have shown that certain HLA alleles and genotypes can influence an individual’s susceptibility to HIV, the progression of the disease, and the response to antiretroviral therapy. However, the role of HLA-DRA gene polymorphisms in HIV infection remains less well understood [6,7,8].This study aims to fill this gap by investigating the polymorphisms of the HLA-DRA gene, particularly focusing on the C and T alleles, in a specific population—individuals infected with HIV in the Samarkand region of Uzbekistan. The Uzbek population presents a unique genetic background, and understanding the distribution of HLA-DRA polymorphisms in this group could provide valuable insights into the genetic factors that influence HIV susceptibility and disease progression.The focus on the C and T alleles is of particular interest because these alleles have been implicated in various immune responses. In other studies, certain alleles of HLA genes have been associated with either a protective effect or an increased risk of various infectious and autoimmune diseases. By analyzing the frequency and distribution of these alleles in HIV-positive individuals compared to a healthy control group, this study seeks to identify potential genetic markers that could be used to predict susceptibility to HIV or to tailor therapeutic strategies based on an individual’s genetic makeup.This study aims to provide a critical step in understanding the interplay between genetic factors and HIV infection, with the potential to contribute significantly to the field of infectious disease genomics and personalized medicine.
2. Materials and Methods
- This study was conducted to explore the distribution of HLA-DRA gene polymorphisms in an HIV-positive cohort from the Uzbek population in the Samarkand region. The study involved a total of 63 HIV-positive individuals and 83 healthy controls. The following sections describe the methodologies employed in the analysis:Study PopulationThe study participants were selected from the Samarkand region of Uzbekistan, comprising individuals diagnosed with HIV and a control group of healthy individuals. The inclusion criteria for the HIV-positive group included a confirmed diagnosis of HIV infection, while the control group consisted of individuals without any known history of HIV or other immunocompromising conditions. The sample size for the HIV-positive group was 63, and for the control group, it was 83.Sample Collection and DNA ExtractionPeripheral blood samples were collected from all participants using standard venipuncture techniques. Genomic DNA was extracted from the blood samples using the [insert kit name and manufacturer, e.g., QIAamp DNA Blood Mini Kit (QIAGEN)], following the manufacturer’s protocol. The quality and concentration of the extracted DNA were assessed using spectrophotometric methods, ensuring the DNA was of sufficient quality for downstream analysis.Genotyping by PCR-RFLPGenotyping of the HLA-DRA gene was performed using the Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) method. This technique was selected due to its well-established sensitivity and specificity for identifying single nucleotide polymorphisms (SNPs), which are critical in studying genetic variability and disease susceptibility. PCR-RFLP enables the differentiation of alleles based on the unique cleavage patterns produced by restriction enzymes when SNPs alter recognition sites. In this case, it was used to distinguish between the C and T alleles within the HLA-DRA gene.Primer Design and PCR AmplificationPrimers were specifically designed to flank and amplify the regions of interest within the HLA-DRA gene that contain the SNPs, ensuring accurate amplification of both C and T alleles. Primer design involved careful consideration of factors such as melting temperature, specificity, and avoidance of secondary structures that could interfere with the reaction.PCR amplification was carried out in a thermocycler under the following conditions:• Initial denaturation at 95°C for 5 minutes to ensure complete melting of the double-stranded DNA.• This was followed by 35 cycles of:ο Denaturation at 95°C for 30 seconds to separate the DNA strands.ο Annealing at the appropriate temperature (usually between 55-60°C) for 30 seconds, allowing the primers to bind specifically to the target sequences.ο Extension at 72°C for 1 minute, during which Taq polymerase synthesized the new DNA strand by adding nucleotides complementary to the template strand.• A final extension step was performed at 72°C for 7 minutes to ensure complete synthesis of all PCR products, especially for any partially extended fragments.Restriction Enzyme Digestion and Gel ElectrophoresisThe amplified PCR products were then subjected to digestion using restriction enzymes specifically chosen for their ability to recognize and cleave the DNA sequence containing either the C or T allele. This step was crucial for distinguishing between the alleles, as the presence of the SNP at the restriction site determines whether the enzyme can cut the DNA. For instance, the enzyme would cleave the DNA at a specific site if the C allele is present, but not if the T allele is present (or vice versa).After digestion, the resulting DNA fragments were separated by size using electrophoresis on a 2% agarose gel. This gel concentration was chosen to optimize resolution for the fragment sizes expected from the digestion. The gel was run in an electrophoresis chamber, where an electric field caused the negatively charged DNA fragments to migrate through the gel matrix. Smaller fragments moved more quickly through the gel, while larger fragments moved more slowly, allowing for clear separation based on size.Visualization of Digestion ProductsOnce electrophoresis was complete, the gel was stained with ethidium bromide, a DNA-intercalating dye that fluoresces under UV light. The stained gel was then viewed using a UV transilluminator to visualize the DNA fragments. The different fragment patterns corresponded to the presence of either the C or T allele, depending on how the DNA was cut by the restriction enzyme. This visualization enabled precise genotyping of the HLA-DRA gene at the SNP of interest.This method provided a reliable and cost-effective means of genotyping, with the advantages of high specificity and reproducibility, making it a valuable tool for the detection of polymorphisms in genetic research.Allele and Genotype Frequency DeterminationThe frequencies of the C and T alleles, as well as the corresponding genotypes (C/C, C/T, T/T), were determined for both the HIV-positive group and the control group. Allelic frequencies were calculated as the proportion of each allele within the total number of alleles observed, while genotypic frequencies were calculated as the proportion of each genotype within the total number of individuals in the respective groups.Statistical AnalysisThe statistical analysis was conducted to evaluate the association between the HLA-DRA polymorphisms and HIV infection. Chi-square (Χ²) tests were employed to assess the significance of differences in allele and genotype frequencies between the HIV-positive and control groups. The odds ratio (OR) and relative risk (RR) were also calculated to quantify the strength of the association between specific alleles/genotypes and the risk of HIV infection.The OR was calculated as the ratio of the odds of having a specific genotype in the HIV-positive group to the odds of having the same genotype in the control group. The RR was calculated as the ratio of the probability of HIV infection in individuals with a specific genotype to the probability of infection in those without that genotype. Confidence intervals (CI) were calculated for both OR and RR to provide an estimate of the precision of these measures.Ethical ConsiderationsAll participants provided informed consent before the collection of blood samples, and the study was conducted following the ethical standards of the Declaration of Helsinki. The study protocol was reviewed and approved by the local ethics committee of Samarkand State Medical Institute.
3. Results
- The results of this study demonstrate significant differences in the distribution of HLA-DRA gene polymorphisms between HIV-positive individuals and healthy controls within the Uzbek population. The analysis revealed a marked difference in the frequencies of the C and T alleles between these two groups. The frequency of the C allele was found to be significantly lower in the HIV-positive group, at 76.98%, compared to 96.38% in the control group. This suggests that the C allele may play a protective role against HIV infection within this population. The relative risk (RR) associated with the C allele was calculated to be 5.24, with a Chi-square (Χ²) value of 32.712, indicating statistical significance with a p-value of 0.021. Conversely, the T allele was more prevalent in the HIV-positive group, with a frequency of 23.01%, compared to just 3.61% in the control group.
|
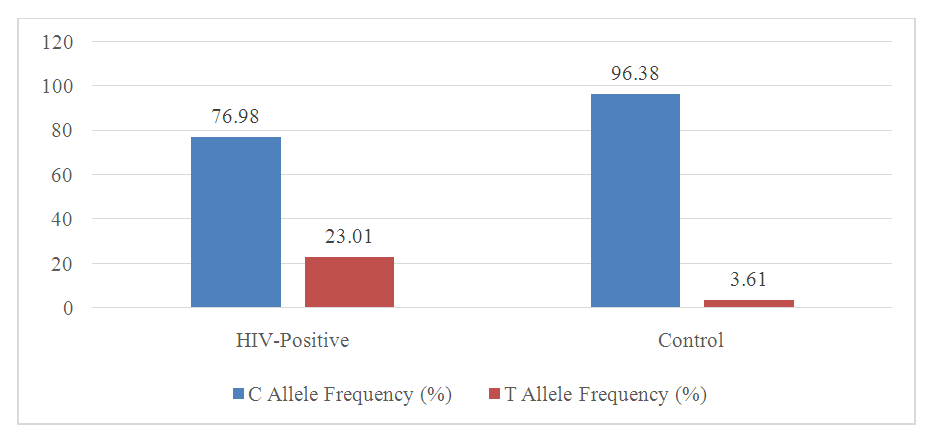 | Figure 1. Allele Frequencies in HIV-Positive and Control Groups |
|
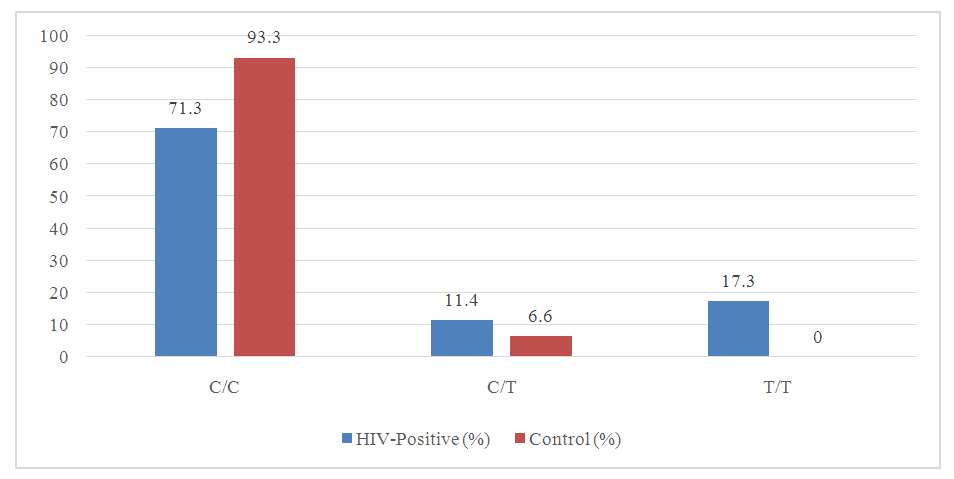 | Figure 2. Genotype Frequencies in HIV-Positive and Control Groups |
4. Conclusions
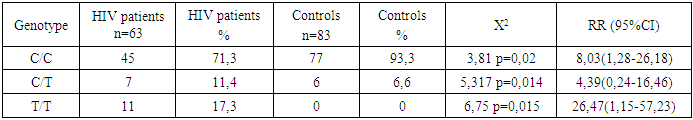
- This study provides significant insights into the genetic factors associated with HIV susceptibility in the Uzbek population by analyzing the polymorphisms of the HLA-DRA gene. The findings reveal that the C and T alleles of the HLA-DRA gene exhibit notable differences in frequency between HIV-positive individuals and healthy controls. Specifically, the C allele, which is more prevalent in the control group, appears to confer a protective effect against HIV infection. In contrast, the T allele is significantly more frequent in the HIV-positive group, suggesting its strong association with increased susceptibility to HIV.Genotype analysis further supports these conclusions, with the homozygous C/C genotype being more common in the control group, reinforcing its protective role. Conversely, the T/T genotype, which is significantly more prevalent in the HIV-positive group, is identified as a major genetic risk factor for HIV infection. These findings underscore the importance of considering genetic predispositions in the epidemiology of HIV and suggest that the HLA-DRA gene, particularly the T allele and T/T genotype, could serve as potential markers for identifying individuals at higher risk of HIV infection.The implications of this study extend beyond the Uzbek population, offering valuable data that could inform global strategies for managing and preventing HIV. By identifying specific genetic markers associated with HIV susceptibility, this research paves the way for the development of personalized medicine approaches. These approaches could tailor prevention and treatment strategies to an individual's genetic profile, potentially improving outcomes and reducing the overall burden of HIV.In conclusion, the significant association between HLA-DRA polymorphisms and HIV infection in this study highlights the critical role of genetic factors in disease susceptibility. Future research should continue to explore these associations in larger and more diverse populations to validate these findings and further elucidate the mechanisms through which these genetic factors influence HIV susceptibility and progression.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML