Shakhnoza Sabirova1, Marguba Zhumaboeva2, Yuliana Assesorova3, Dilfuza Matkarimova4, Timur Alimov5, Kodirjon Boboev5
1Research Grant, Republican Specialized Scientific and Practical Medical Center of Hematology (RSSPMCH) MoH RUz, Tashkent, Uzbekistan
2Hematology Department, Multidisciplinary Medical Center of Khorezm region, Khorezm, Uzbekistan
3Laboratory of Molecular Genetics, Cytogenetics and FISH, Republican Specialized Scientific and Practical Medical Center of Hematology (RSSPMCH) MoH RUz, Tashkent, Uzbekistan
4Department of Hematology, Transfusion and Laboratory Science, Tashkent Medical Academy, Tashkent, Uzbekistan
5Medical Genetics Laboratory, Republican Specialized Scientific and Practical Medical Center of Hematology (RSSPMCH) MoH RUz, Tashkent, Uzbekistan
Correspondence to: Timur Alimov, Medical Genetics Laboratory, Republican Specialized Scientific and Practical Medical Center of Hematology (RSSPMCH) MoH RUz, Tashkent, Uzbekistan.
| Email: |  |
Copyright © 2024 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Abstract
Introduction. Chronic myeloid leukemia (CML) is a disease of the hematopoietic system of tumor origin, in the pathogenesis of which lies a translocation between chromosomes 9 and 22. Aim. Study and analysis of cytogenetic and molecular genetic findings in patients with chronic myeloid leukemia, and development of a new diagnostic model. Main findings. The material for the study was 204 patients diagnosed with CML. Clinical, laboratory and molecular studies were performed, which revealed: out of 204 total patients, 5 patients had additional chromosomal abnormalities (DCA) in standard cytogenetic study, and molecular genetic studies showed a high level of correlation of positive BCR::ABL p210 transcripts (189 vs. 191, range from 0.009 to 100%; at r=0.979) in 204 patients with CML.
Keywords:
Gene, Chronic myeloleukemia, Standard cytogenetics, Polymerase chain reaction, Analysis, Patients
Cite this paper: Shakhnoza Sabirova, Marguba Zhumaboeva, Yuliana Assesorova, Dilfuza Matkarimova, Timur Alimov, Kodirjon Boboev, Cytogenetic and Molecular Genetic Aspects of Chronic Myeloid Leukemia in Uzbekistan, American Journal of Medicine and Medical Sciences, Vol. 14 No. 6, 2024, pp. 1635-1639. doi: 10.5923/j.ajmms.20241406.35.
1. Introduction
Chronic myeloid leukemia (CML) is a disease of the hematopoietic system, which is of tumor origin, resulting from qualitative, quantitative inferiority with impaired function of white blood cells – leukocytes, resulting in an increase in some of their forms in the peripheral blood [1,5].Peter Cullen first described a patient with acute inflammation of the spleen and “milky blood” in 1811. Sometime later in 1845, when cell staining techniques were not yet in use but microscopes were already in use, Scottish pathologist John Bennett described in his scientific papers tissues of enlarged spleen and liver obtained from two patients who were fatal “from blood poisoning.” German pathologist Rudolf Virchow literally in 1,5 months published a similar picture, and 2 years later he, having found almost such a picture, for the first time gave a name to the disease of the supposed disease as - “splenic leukemia”. With the advent of methods for staining cells for microscopic examination in 1880, scientists were able to examine cells with this pathology in detail. In 1950-1960, American researchers P. Nowell and D. Hungerford on the basis of cytogenetic studies found that in all patients with CML one of the chromosomes was shortened, and moreover that they had a clonal character, which from a single mutated cell develops a pathological process [23].Later, Janet D. Rowley determined that in the pathogenesis of CML plays not only a shortened chromosome, but also there is a translocation between the 9 and 22 chromosomes, the 9 chromosome that contains the oncogene ABL is joined with the BCR gene located on the 22nd chromosome, and as a result, the so-called chimeric hybrid BCR::ABL gene is formed. These findings have made a huge contribution to the understanding that cytogenetic studies are a mandatory method for determining the forms of leukemia, as they play a major role in studying the pathogenesis of the disease [6,14].Advances in the study of disease pathogenesis (CML) have made it possible to meaningfully change the quality and length of life of patients with CML and the question of how to completely cure patients with this pathology is currently being considered worldwide [2,4,20,21].The disease ranks 3rd among all blood cancers, accounting for about 20-25% of cases in North Africa, Europe [13], and in Asian countries like India, Japan accounting for about 30-35% of cases comes in 2nd place and can be diagnosed with an incidence rate of about 1-1.5 per 100,000 people in all countries of the world [22,26,27].
2. Main Body
2.1. Purpose of the Study
Study and analysis of cytogenetic and molecular genetic findings in patients with chronic myeloid leukemia, and development of a new diagnostic model.
2.2. Material and Methods of Investigation
Own data: 204 patients with chronic myeloleukemia who were initially admitted (n=134) to the consulting and diagnostic department of the Republican Specialized Scientific and Practical Medical Center of Hematology (RSSPMCH) of MoH RUz for diagnosis and subsequent treatment and registered with the same diagnosis in RSSPMCH (n=70) were selected for the scientific study. The diagnosis was verified on the basis of clinical, laboratory (general blood analysis, myelogram with cytochemistry, cytogenetics and FISH, molecular genetic studies) data.The incidence ratio by gender was 1.4:1 men vs. women. The median age was 50 years (from 16 to 74), the peak incidence was 50-59 years, but the proportion of young patients under 44 years of age was significant: up to 46% (Fig.1,2).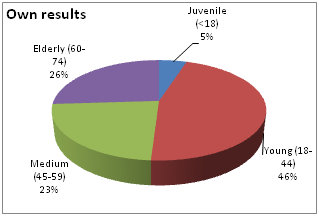 | Figure 1. Age differences in the incidence of the disease |
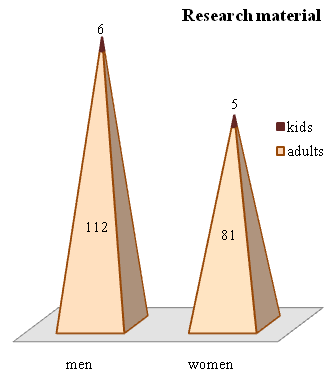 | Figure 2. Distribution of patients by gender and age category (n=204) |
Molecular genetic analysis for BCR::ABL p210 was performed among the whole study sample of CML patients.The studies were performed on mRNA preparations taken from peripheral blood cells and bone marrow of 204 patients diagnosed with CML. The level of chimeric BCR::ABL p210 gene was determined by real-time PCR on a Rotor-Gene Q thermocycler. To evaluate the dynamics of changes in BCR::ABL gene expression, the International Scale (IS) recommended by the Expert Group of European Leukemia Net (ELN) was used, while the expression of the control ABL gene was considered as 100% [15,18,19,25].Statistical processing of data was performed using Excel software (Biostatistics 4.03), which included determination of median, maximum and minimum values, correlation coefficient. The data were processed in Statistica 5.0 for Windows. The results were considered reliable at p<0.05.
2.3. Results and Discussion
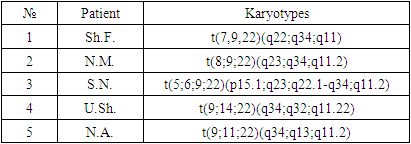
Standard cytogenetic studyOne of the main genetic methods of investigation of CML patients is the standard cytogenetic study (SCI). This method allows to evaluate the whole chromosomal set of leukemia cells and to detect not only Philadelphia (Ph) chromosome, which is a marker cytogenetic manifestation of pathogenetic translocation t(9;22)(q34;q11.2), but also to estimate the number and structural integrity of other chromosomes.While the marker translocation t(9;22) and major additional rearrangements [12] can be detected by other methods (FISH, PCR, PCR-RT), information about rare and therefore poorly studied chromosomal rearrangements can only be obtained by analyzing GTG-stained metaphase chromosomes. For this reason, in the European Leukemia Net (ELN) recommendations [7], as well as in the Cytogenetic Recommendations of the pan-European system of quality assessment of constitutional and acquired cytogenetic studies developed by the European Cytogeneticists Association (E.C.A.) [11], the analysis of metaphase chromosomes (G-banding) is included in the list of mandatory elements of the algorithm of diagnostic examination of CML patients and monitoring of the course of the disease.Along with this, the method of fluorescent in situ hybridization FISH is currently one of the most important laboratory diagnostic methods for many diseases, including CML. The analysis allows to detect in the genetic material of blood cells and bone marrow mutant gene, which plays an important role in the development of the disease at the molecular level. Using this method it is possible to detect a pathological clone even in the absence of dividing cells, the superiority of the study is that the method can detect the pathological process, using even peripheral blood samples. To detect BCR::ABL1 fusion gene, DNA probes labeled with different fluorochromes are used, and the chromosome rearrangement can be detected in 95% of first-time patients with clinical diagnosis of CML [26].Cytogenetic and FISH studies at the stage of diagnosis allowed us to identify out of 204 (93% Ph+) patients studied, tumor cell clones with variant translocations t(V;9;22) formed with the participation of three chromosomes in 2 (0.9%) cases and additional chromosomal aberrations (ACA) in Ph-positive cells in 3 (1.4%) patients (Table 1).Table 1. Bone marrow karyotypes in newly diagnosed CML patients with variant translocations t(V,9,22) and additional chromosomal abnormalities in Ph-positive cells
 |
| |
|
In addition to the pathogenetic translocation t(9;22)(q34;q11.2), the cytogenetic manifestations of which are derivatives of chromosome der22 (Ph-chromosome) and der9, karyotyping using G-banding technology allows to detect atypical variants of this rearrangement, as well as additional chromosomal abnormalities that arise with increasing genetic instability due to the inability of leukemia cells to repair new DNA damage and apoptosis.Atypical translocation variants involving chromosomes 9 and 22 (variant translocations) can occur in 5-10% of patients [24].The peculiarity of these rearrangements is the involvement of fragments of other chromosomes in the recombination of chromosomes 9 and 22. In this case, 3 or more chromosomes can become participants of variant translocation. However, at any number of participants and loci of breaks at variant translocation, the chimeric oncogene BCR::ABL1 pathogenetic for CML is retained on der22 (Fig. 3). Additional chromosomal anomalies (ACA) arising as a consequence of genetic instability are independent mutations and are not associated with derived chromosomes 9 and 22, in any case, the exchange of fragments does not affect the 9q34 and 22q11.2 break loci involved in the formation of BCR::ABL1.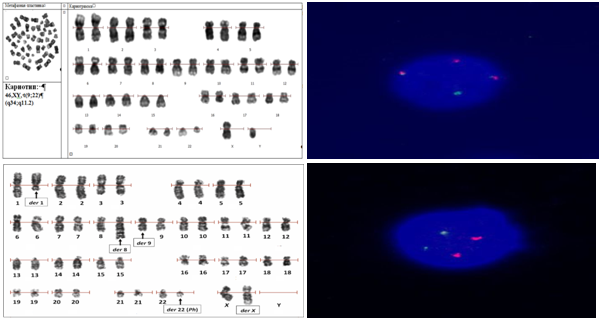 | Figure 3. Cariogram: a. 46,ХY, t(9;22) (q34;q11.2); b. Translocation of BCR::ABL1 (on the Ph chromosome) and ABL1::BCR (altered chromosome 9) c. Variant translocation in CML with additional chromosomal rearrangement; d. Norm. The two red signals ABL1 and the two green signals BCR lie separately. There are no fusion signals (own data) |
Molecular genetic research.Polymerase chain reaction (PCR) is a molecular-genetic method that is used both for diagnostic purposes and for monitoring therapy in CML [8,10,27]. The sensitivity of this method is so high that it is possible to detect a single cell with specific DNA from about 105 cells.Fundamentals of the method:a) total RNA extractions from peripheral and venous blood cells, sternum punctate cells; b) reverse transcription reactions; c) amplification with “real-time” detection with two oligonucleotide mixtures.Own results of molecular genetic studiesWhen studying the expression of BCR::ABL p210 gene, primers to standard variants of BCR::ABL p210 gene were used. The studies were performed according to the method of BCR::ABL1 gene mRNA detection in clinical material.The results of the analysis were interpreted based on the expression level of the chimeric oncogene BCR::ABL in relation to the expression of the control gene ABL1 (Fig. 4). The threshold value for a positive result of the analysis: the expression level was >0.01%; for a negative result: the expression level was <0.01%.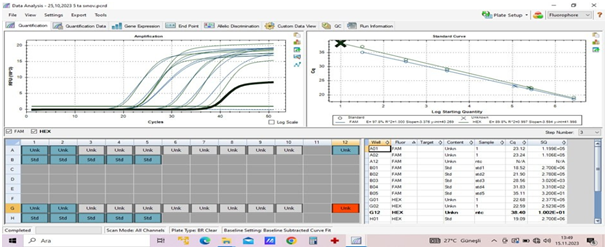 | Figure 4. BCR::ABL p210 gene expression (Own data) |
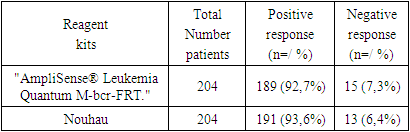
The analysis of the evaluation of DNA samples from the manufacturer of Russian “AmpliSense® Leukemia Quantum M-bcr-FRT” of CML diagnostics intended for PCR assay showed the following results. Thus, for quantitative evaluation of BCR::ABL p210 transcripts in 204 CML patients, expression was analyzed by RT-PCR method (Table 2).Table 2. Quantification of BCR::ABL p210 transcripts
 |
| |
|
Along with this, we decided to make changes to the instructions from the manufacturer of AmpliSense (Russia), in order to develop our own new method. Since, in the future, the introduction of a new method in the study of BCR::ABL p210 oncogene expression will allow hematologists to monitor the condition of patients in the dynamics of treatment, including after stem cell transplantation, to study the minimal residual disease and predict the occurrence of relapses.According to the manufacturers' instructions for the initial lysis step, the lysate should have been incubated in the refrigerator for 10 minutes, but we decided to increase the incubation time by another 5 minutes, because when we set it to 10 minutes the results were slightly lower than expected. The next improved step was the 4th extraction step, when the leukocyte count is 2x106 we should have used 350 µl of RLT buffer as indicated in the instructions, and when the leukocyte count is between 2x106 and 1x107 we should have used 600 µl of buffer. However, this was difficult to implement because the leukocyte counts are not the same in patients diagnosed with CML before and after chemotherapy, so we used RLT buffer according to the amount of pure leukocyte extract.After making these changes, the amount of extracted RNA was sufficient and pure for the next steps of the study. When the amount of RNA in the nanodrop with concentration from 50 to 1500 µg/mol was measured, a high purity coefficient was obtained and the result was reliable 260/280=2 23/260=1.80.Of note, 7.3% and 6.4% of BCR::ABL p210 samples were not detected by both methods, the detected differences between the AmpliSense® Leukemia Quantum M-bcr-FRT and Nou hau kit did not reach a statistically significant level (χ2=0.4; p=0.5; OR=1.2; 95%CI:0.58-2.78). The observed high level of correlation of BCR::ABL p210 transcript positivity in (189 vs. 191, range 0.009 to 100%; at r=0.979) 204 patients with CML demonstrates the concordance of the results obtained using comparative methods in detecting bcr/abl p210 transcript. In addition, a positive correlation was found between the percentage of Ph-positive tumor clones and the expression level of BCR::ABL p210. The Spearman's rank correlation coefficient was 0.824 (p < 0.001). These data allow us to use the level of bcr/abl p210 molecular expression to consider the tumor load and, if necessary, to make an approximate assessment of the cytogenetic response to the current therapy, which is especially important when there will be no possibility to perform SCI [9,16,17].After that, we monitored treatment in 70 patients registered with CML. For this purpose, molecular genetic study of BCR::ABL p210 gene expression in peripheral blood cells was repeated after 6 months on average. The study showed that in the group of patients (n=70) registered with the diagnosis of CML, 14.3% of patients had a large molecular response (BMR) and the rest of patients with positive response dynamics during imatinib therapy (86.6%) showed a decrease in the level of BCR::ABL p210 gene expression by an average of 2 lg relative to the previous result.Achievement of BMO, defined as BCR::ABL gene expression less than 0.1% is the most important criterion of molecular response to CML therapy, in our case this drug was Glivec (1st generation tyrosine kinase inhibitor).According to the literature of world researchers and according to ELN recommendations, a period of at least 18 months is required to achieve a higher BMO result [3,8], and our studies lasted less than this period, due to which 86.6% of patients currently have insignificantly higher expression of the chimeric gene. Moreover, as we mentioned above, we have developed a new modified method to determine the expression of BCR::ABL p210 gene, in the future it will allow patients and hematologists to clarify the pathology more in the early stages of the disease and create convenience and accessibility in economic terms.
3. Conclusions
In conclusion, we can state that the application of all the above mentioned diagnostic and treatment monitoring methods is reasonable for the study and control of the tumor clone, expression level and for the assessment of the mutational status of the BCR::ABL chimeric oncogene. Molecular-genetic monitoring in the future will allow to adequately assess the effectiveness of treatment of this disease and, if necessary, to take timely measures to correct the therapy [23,24,25].The study is ongoing, as the next stage of work is to clarify the relationship and intersectionality of our studies, that is, to analyze the correlation between all these parameters listed below: • age and gender of patients with this pathology. • with the results of laboratory tests on the other hand. • duration of the period from the moment of clinical diagnosis of CML to the beginning of therapy with Imatinib drug.• duration of Imatinib therapy. • time to achieve a complete cytogenetic response. • timeframe for achieving a complete molecular response.
References
| [1] | Ahmed A. A. et al. Chronic Myeloid Leukemia: A retrospective study of clinical and pathological features // Bionatura. – 2022. – Vol. 7. – №. 3. – P. 41. doi. 10.21931/RB/2022.07.03.41. |
| [2] | Algahtani F.H., Alqahtany F.S. Evaluation and characterisation of Chronic myeloid leukemia and various treatments in Saudi Arabia: A retrospective study // Journal of infection and public health. – 2020. – Vol. 13. – №. 2. – P. 295-298. |
| [3] | Baccarani M. et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013 // Blood, The Journal of the American Society of Hematology. – 2013. – Vol. 122. – №. 6. – P. 872-884. |
| [4] | Berman E., Shah N.P., Altman J.K. et al. CML and the WHO: Why? Journal of Clinical Oncology Volume 42, Number 9 З. 984-986 https://doi.org/10.1200/JCO.23.0168. |
| [5] | Bi X. et al. The first case of six-way complex translocation of t (4; 7; 9; 22; 8; 14) in a patient with chronic myeloid leukemia // Journal of Hematopathology. – 2024. – P. 1-5. doi: 10.1007/s12308-024-00577-8. |
| [6] | Cortes J. E. et al. Ponatinib efficacy and safety in Philadelphia chromosome–positive leukemia: final 5-year results of the phase 2 PACE trial // Blood, The Journal of the American Society of Hematology. – 2018. – Vol. 132. – №. 4. – P. 393-404. |
| [7] | Cortes J.E. et al. Ponatinib efficacy and safety in Philadelphia chromosome–positive leukemia: final 5-year results of the phase 2 PACE trial // Blood, The Journal of the American Society of Hematology. – 2018. – Vol. 132. – №. 4. – P. 393-404. |
| [8] | Cross NCP, Ernst T, Branford S, et al. European Leukemia Net laboratory recommendations for the diagnosis and management of chronic myeloid leukemia. Leukemia. 2023 Nov; 37(11): 2150-2167. doi: 10.1038/s41375-023-02048-y. |
| [9] | Dakkoune M. et al. Imatinib dans le traitement de la leucémie myéloide chronique au Maroc // Bulletin du Cancer. – 2020. – Vol. 107. – №. 9. – P. 861-866. https://doi.org/10.1016/j.bulcan.2020.05.013. |
| [10] | Dewald G.W. et al. Highly sensitive fluorescence in situ hybridization method to detect double BCR/ABL fusion and monitor response to therapy in chronic myeloid leukemia // Blood. American Society of Hematology, 1998. – Vol. 91. – № 9. – P. 3357–3365. |
| [11] | Guru Murthy G. S. How I Manage Patients with Chronic Myeloid Leukemia (CML): Perspectives from Clinical Practice // Blood and Lymphatic Cancer: Targets and Therapy. – 2022. – P. 1-6. |
| [12] | Hastings R.J. et al. Cytogenetic guidelines and quality assurance: a common European framework for quality assessment for constitutional and acquired cytogenetic investigations // European Journal of Human Genetics. – 2007. – Vol. 15. – №. 5. – P. 525-527. |
| [13] | Henke O. et al. Early molecular response in East African Philadelphia chromosome-positive chronic myeloid leukaemia patients treated with Imatinib and barriers to access treatment //ecancermedicalscience. – 2020. – Vol. 14. – P. 1-11. |
| [14] | Hill A. W., Perry M. J., Levine P. H. Agricultural practices and age of chronic myeloid leukemia diagnosis in India // Clinical Epidemiology and Global Health. – 2018. – Vol. 6. – №. 2. – P. 56-60. |
| [15] | Hochhaus A, Baccarani M, Silver RT, et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia. 2020 Apr; – Vol. 34. – №4. – P. 966-984. doi: 10.1038/s41375-020-0776-2. |
| [16] | Hochhaus A. et al. Long-Term Outcomes of Imatinib Treatment for Chronic Myeloid Leukemia // N. Engl. J. Med. 2017. Vol. 376. – № 10. – P. 917–927. |
| [17] | Hoffmann V.S. et al. Treatment and outcome of 2904 CML patients from the EUTOS population-based registry // Leukemia. Nature Publishing Group, 2017. – Vol. 31. – № 3. – P. 593–601. |
| [18] | http://www.leukemianet.org/content/leukemias/cml/cmlscore/index_eng.html |
| [19] | Kumar A. et al. Higher incidence of co-expression of BCR-ABL fusion transcripts in an Eastern Indian population // Egyptian Journal of Medical Human Genetics. – 2023. – – Vol. 24. – №. 1. – P. 56. |
| [20] | Lin Q. et al. Global, regional, and national burden of chronic myeloid leukemia, 1990–2017: a systematic analysis for the global burden of disease study 2017 // Frontiers in Oncology. – 2020. – Vol. 10. – P. 580759. doi: 10.3389/fonc.2020.580759. |
| [21] | Mughal T. I. et al. Chronic myeloid leukemia: reminiscences and dreams // Haematologica. – 2016. – Т. 101. – №. 5. – С. 541. |
| [22] | Ning L. et al. Trends in disease burden of chronic myeloid leukemia at the global, regional, and national levels: a population-based epidemiologic study // Experimental hematology & oncology. – 2020. – Т. 9. – С. 1-14. |
| [23] | Sokal J.E. et al. Prognostic discrimination in “good-risk” chronic granulocytic leukemia // Blood. 1984. Vol. 63, № 4. P. 789–799. |
| [24] | Son H. J., Lee J. H. Novel Four-Way Variant Translocation, t (1; 9; 22; 16)(q21; q34; q11. 2; q24), in a Patient with Chronic Myeloid Leukemia // Diagnostics. – 2024. – Т. 14. – №. 3. – С. 303. |
| [25] | Tariq I. Mughal, Jerald P. Radich et al. Daley Chronic myeloid leukemia: reminiscences and dreams, J. Haematologica 2016, – Vol.101. – №5. – P. 541-558. |
| [26] | Ursan I.D. et al. Emergence of BCR-ABL Kinase Domain Mutations Associated with Newly Diagnosed Chronic Myeloid Leukemia: A Meta-Analysis of Clinical Trials of Tyrosine Kinase Inhibitors // J. Manag. Care Spec. Pharm. 2015. Vol. 21. – № 2. – P. 114–122. |
| [27] | Zaker E. et al. The importance of personalized medicine in chronic myeloid leukemia management: a narrative review // Egyptian Journal of Medical Human Genetics. – 2023. – Vol. 24. – №. 1. – P. 31. |





 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML

