-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2023; 13(11): 1672-1678
doi:10.5923/j.ajmms.20231311.18
Received: Oct. 19, 2023; Accepted: Nov. 5, 2023; Published: Nov. 9, 2023

Molecular Genetic Analysis of the Correlation of Allelic Polymorphisms of Xenobiotic Genes with the Development of Nonsyndromic Cleft Lip and/or Palate
P. B. Gulmukhamedov1, J. A. Rizaev2, N. L. Khabilov3, K. T. Boboev4
1Doctoral student, PhD, Samarkand State Medical University, Uzbekistan
2Rector, Doctor of Medical Sciences, Prof. Samarkand State Medical University, Uzbekistan
3Head of Department in Hospital Orthopedic Dentistry, MD, Prof. Tashkent State Dental Institute, Uzbekistan
4Republican Specialized Scientific-Practical Medical Center for Hematology of the Ministry of Health of the Republic of Uzbekistan
Copyright © 2023 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

A molecular genetic analysis of the relationship of allelic polymorphisms of the xenobiotic system genes (MDR1 (C1236T) and MDR1 (C3435T)) with the development of nonsyndromic congenital malformations of the maxillary region in Uzbekistan was carried out. According to the results of the study, a possible contribution of the C/T genotype of the polymorphic gene MDR1 (C1236T) to an increased risk of the formation of an isolated cleft lip (Q36) was established. Whereas with respect to polymorphic loci of polymorphism of the MDR1 gene (C3435T), the absence of their participation in the processes of formation of the ERW in Uzbekistan was established.
Keywords: Congenital malformations of the maxillary region, MDR1 (C1236T), MDR1 (C3435T), Carriers, Isolated cleft palate (Q35), Isolated cleft lip (Q36), Combined cleft palate and lip (Q37)
Cite this paper: P. B. Gulmukhamedov, J. A. Rizaev, N. L. Khabilov, K. T. Boboev, Molecular Genetic Analysis of the Correlation of Allelic Polymorphisms of Xenobiotic Genes with the Development of Nonsyndromic Cleft Lip and/or Palate, American Journal of Medicine and Medical Sciences, Vol. 13 No. 11, 2023, pp. 1672-1678. doi: 10.5923/j.ajmms.20231311.18.
1. Relevance
- Over the past two decades, due to the widespread introduction of high-tech research methods (molecular genetic analysis) into science, significant progress has been made in understanding the mechanisms of the formation of a large number of diseases, including congenital malformations of the maxillary facial region (NSCL/P) [1,2,3,4,9]. Many studies have been conducted, the results of which allowed us to study the contribution of various polymorphic genes in a very wide list of diseases [5,6,8,10,13]. In particular, few studies have been conducted regarding the study of the contribution of these genes to the initiation of pathological processes that contribute to the onset of HPV [7,9,14,15].Of particular interest among a large group of genes of the xenobiotic system are various polymorphisms of the MDR1 gene [6,13]. It is reported that the MDR1 gene, located on chromosome 7, is expressed in the plasma membranes of cells and organs, encoding cellular transmembrane P-glycoprotein, which removes a wide range of xenobiotic compounds from cells [1,2,9]. The most common polymorphic variants of the MDR1 gene are variants MDR1 (C1236T) and MDR1 (C3435T) [3,4].Changes in the structure of the MDR1 gene are accompanied by disturbances in its activity, which is fraught with the launch of complex pathological processes that form the basis for the development of NSCL/P [11,12,16,17,18], which served as the basis for conducting studies to assess the contribution of MDR1 polymorphism (C3435T) to the formation of HPV in Uzbekistan.
2. Material and Methods
- The study involved 105 children (average age 6.5±1.8 years) with NSCL/P (the main group of NSCL/P) living in the territory of the Republic of Uzbekistan, who were monitored at the clinic of the Tashkent State Dental Institute in the period from 2019 to 2022 In accordance with the international classification of diseases of the 10th revision (ICD 10) all children with NSCL/P (n=105) were divided into three groups depending on nosology: Q35 (n=35) – children with cleft palate; Q36 (n=33) – children with cleft lip; Q37 (n=37) – children with cleft palate and lip. The compared control group consisted of 103 healthy children without a history of congenital malformations, comparable in place of residence, age and gender with the main group of children with NSCL/P. Molecular genetic studies of polymorphic loci of the MDR1 (C1236T) and MDR1 (C3435T) genes were carried out in the laboratory of Molecular Genetics, Cytogenetics and FISH republic specialized scientific practical medical center of hematology (RSSPMCH, Republic of Uzbekistan, Tashkent). In accordance with the generally accepted methodology, DNA was isolated from blood leukocytes and the polymorphisms MDR1 (C1236T) and MDR1 (C3435T) (Rotor Gene Q, Quagen, Germany) were studied. The results were processed using the statistical program "OpenEpi 2009, Version 9.2".
3. Results and Discussion
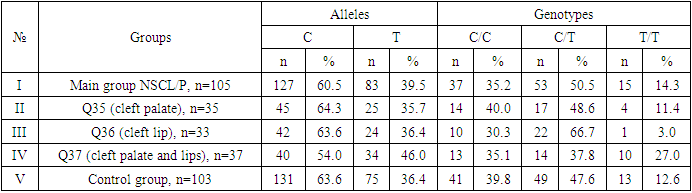
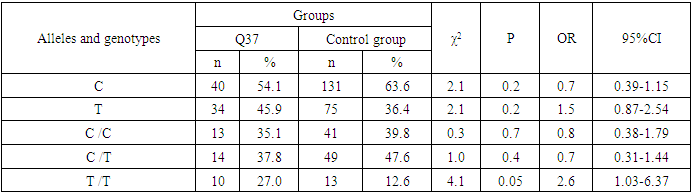
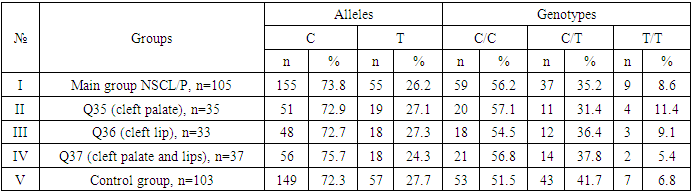
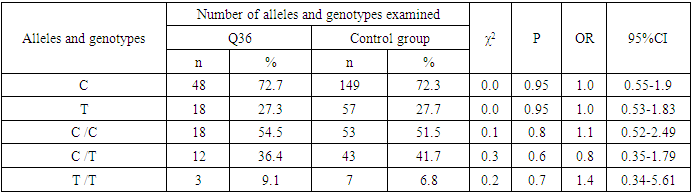
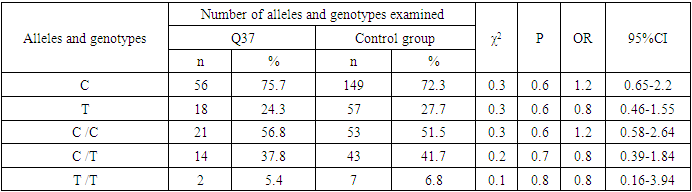
- During the analysis of the distribution of expected and observed genotype frequencies of polymorphic genes MDR1 (C1236T) and MDR1 (C3435T) in groups of patients with NSCL/P and healthy, their compliance with the canonical distribution of the Hardy-Weinberg equilibrium was established (HWE) (р>0.05). The distribution of the polymorphic gene MDR1 (C1236T) in the main group of patients with NSCL/P (n=105) and the group of healthy (n=103) is presented in Table 1.
|
|
|
|
|
|
|
|
|
|
4. Conclusions
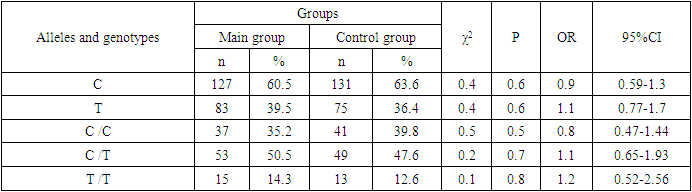
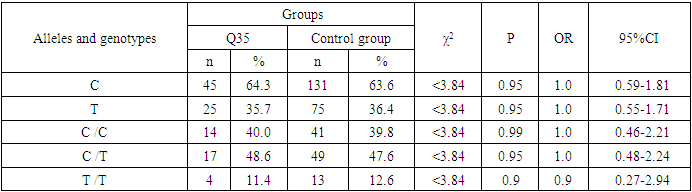
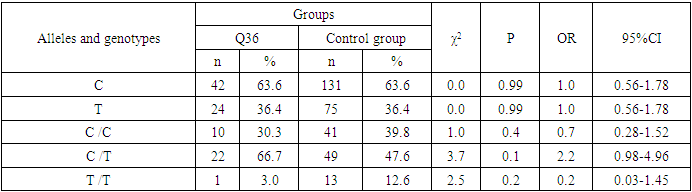
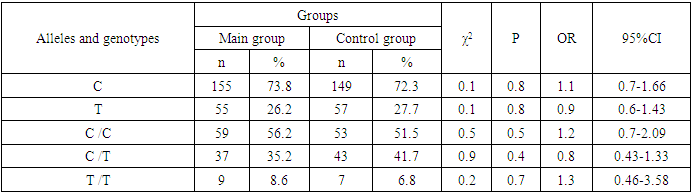
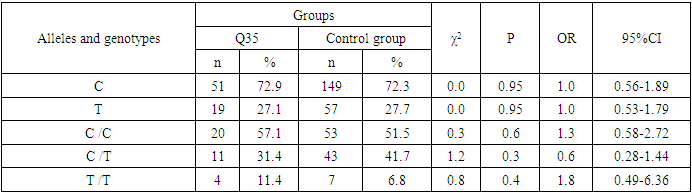
- Analyzing the results obtained, according to the study of the features of the occurrence of alleles and genotypes of the polymorphism of the MDR1 gene (C1236T) in the main group of patients with HPV and with isolated cleft palate (Q35) compared with healthy ones, there were no statistically significant differences in the proportion of the frequency distribution of alleles (for the allele T- χ2=0.4; P=0.6; OR=1.1 and χ2<3.84; P=0.95; OR=1.0, respectively) and genotypes (for the C/T genotype - χ2=0.2; P=0.7; OR=1.1 and χ2<3.84; P=0.95; OR=1.0, respectively; for the genotype T/T - χ2=0.1; P=0.8; OR=1.2 and χ2<3.84; P=0.9; OR=0.9, respectively) according to the variant of the MDR1 gene (C1236T), which proves the absence of its role as a genetic marker predisposing to an increased risk of HPV formation in the main group and isolated cleft palate (Q35).However, between groups with isolated cleft lip (Q36) and healthy in the absence of significant differences in the carrier of the T allele (χ2<3.84; P=0.99; OR=1.0) the presence of a tendency to increase the occurrence of the C/T genotype (χ2=3.7; P=0.1; OR=2.2) among the patients of this group. In turn, this shows the possible contribution of the C/T genotype by the polymorphic gene MDR1 (C1236T) to an increased risk of the formation of an isolated cleft lip. In addition to these facts, statistically significant differences were found between the groups with Q37 and the control in the carrier of the minor T/T genotype by 2.6 times (χ2=4.1; P=0.05), which proves the association of the T/T genotype with an increased risk of Q37 formation. At the same time, between the groups of patients with Q35 and Q36, there were also no statistically significant differences when comparing the frequency of the T allele (χ2<3.84; P=0.95; OR=1.0), the heterozygous C/T genotype (χ2=2.3; P=0.2; OR=0.5) and mutant genotype T/T (χ2=1.8; P=0.2; OR=4.1) by polymorphism of the MDR1 (C1236T) gene among patients with Q35.In addition, differences in the carrier of alleles and genotypes according to the polymorphism of the MDR1 (C1236T) gene between the groups of patients with Q35 and Q37 were also characterized by the absence of statistically significant differences in the proportion of the carrier of the minor allele T (χ2=1.6; P=0.3; OR=0.7), genotypes C/C (χ2=0.2; P=0.7; OR=1.2), S/T (χ2=0.8; P=0.4; OR=1.6) and T/T (χ2=2.8; P=0.1; OR=0.3) among patients with Q35.Comparing the proportions of carriers of alleles and genotypes of the MDR1 (C1236T) gene polymorphism between groups of patients with Q36 and Q37, a statistically significant association with an increased risk of Q37 was found among carriers of heterozygous S/T (χ2=5.8; P=0.025) and mutant T/T (χ2=7.6; P=0.01) genotypes.The results of a comparative assessment of the differences in the frequency distribution of alleles and genotypes for the polymorphic gene MDR1 (C3435T) among the studied groups of patients with HPV and healthy, were characterized by the absence of their statistical significance. Thus, in the main group of patients with HPV, compared with the control, differences for the minor allele T (χ2=0.1; P=0.8) and a heterozygous variant of the S/T genotype (χ2=0.9; P=0.4) did not reach one, while the proportion of mutant genotype T/T among patients was 1.3 times higher (χ2=0.2; P=0.7). In the groups of patients with Q35, Q36 and Q37, compared with the control, the absence of statistical significance of the revealed differences was found both in the distribution of the minor T allele (Q35- χ2<3.84; P=0.95; OR=1.0; Q36 - χ2<3.84; P=0.95; OR=1.0; Q37 - χ2<0.3; P=0.6; OR=0.8), as well as C/T genotypes (Q35 - χ2<3.84; P=0.95; OR=1.0; Q36 - χ2=0.3; P=0.6; OR=0.8; Q37 - χ2=0.3; P=0.6; OR=0.8) and T/T (Q35 - χ2=0.8; P=0.4; OR=1.8; Q36 - χ2<3.84; P=0.7; OR=1.4; Q37 - χ2<3.84; P=0.8; OR=0.8).Statistical significance was also not found when assessing the frequencies of the distribution of alleles and genotypes of the polymorphism of the MDR1 gene (C3435T) between groups of patients with Q35 and Q36 (for T - χ2<3.84; P=0.99; OR=1.0; for genotype C/T - χ2=0.2; P=0.7; OR=0.8 and for the mutant genotype T/T – χ2=0.1; P=0.8; OR=1.3); Q35 and Q37 (for T - χ2=0.1; P=0.7; OR=1.2; for the C/T genotype - χ2=0.3; P=0.6; OR=0.8 and for the mutant genotype T/T – χ2=0.9; P=0.4; OR=2.3) as well as Q36 and Q37 (for T - χ2=0.2; P=0.7; OR=1.2; for genotype C/T - χ2<3.84; P=0.9; OR=0.8 and for the mutant genotype T/T – χ2=0.4; P=0.6; OR=1.8). In this regard, based on the results of the study, we found that the polymorphism MDR1 (C1236T) can increase the likelihood of the formation of Q36, and the polymorphism of the gene MDR1 (C3435T) does not participate in the formation of ERW in Uzbekistan.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML