-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2023; 13(10): 1580-1583
doi:10.5923/j.ajmms.20231310.44
Received: Oct. 9, 2023; Accepted: Oct. 21, 2023; Published: Oct. 25, 2023

Detection of Allelic Variants and Association of Filaggrin Gene Polymorphism Genotypes in Dermatomycoses Among Children in the Uzbek Population
Sh. Z. Mavlyanova, N. Dj. Ikramova, G. R. Ibragimova, I. A. Samarkhodjaeva, Sh. K. Yuldasheva
Republican Specialized Scientific and Practical Medical Center for Dermatology and Venereology, Tashkent, Uzbekistan
Copyright © 2023 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

The pathogenesis of fungal skin diseases highlights the importance of the skin's barrier function - the genes encoding epidermal proteins responsible for forming the epidermal barrier. This study aimed to investigate allelic variants and associations of the 2282del4 polymorphism genotypes in the FLG gene among children with fungal skin and scalp diseases in the Uzbek population. Materials and Methods: We examined 16 children aged 1 to 18 with fungal diseases caused by dermatophytes. Clinical, immunological, molecular genetics and statistical analyses were conducted for all children. Results: Molecular genetic analysis of the frequency distribution of the 2282del4 polymorphism genotypes in the FLG gene revealed an increased heterozygous variant of the FLG gene in the primary group of children with fungal diseases in 31.3% (5/16) of cases, whereas it was not detected in the control group (χ2=10.52, P<0.005; OR=29.2; 95% CI 1.49 - 570.6). The mutant homozygous variant of the 2282del4 polymorphism in the FLG gene was not detected in both groups (χ2=10.52, P<0.005; OR=1.8; 95% CI 0.04 - 97.5). Conclusion: The heterozygous genotype variant of the 2282del4 polymorphism in the FLG gene is a significant marker of an increased risk of developing fungal diseases caused by Trichophyton and Microsporum dermatophytes among children in the Uzbek population (χ2=10.52, P<0.005; OR=29.2; 95% CI 1.49 - 570.6). The functionally favorable genotype of the 2282del4 polymorphism in the FLG gene is a significant protective marker against pathology (χ2=10.5, P<0.005; OR=0.03; 95% CI 0.0-0.67).
Keywords: Dermatomycoses, Trichophytosis, Microsporia, Filaggrin gene, 2282del4 FLG gene polymorphism, Genetics
Cite this paper: Sh. Z. Mavlyanova, N. Dj. Ikramova, G. R. Ibragimova, I. A. Samarkhodjaeva, Sh. K. Yuldasheva, Detection of Allelic Variants and Association of Filaggrin Gene Polymorphism Genotypes in Dermatomycoses Among Children in the Uzbek Population, American Journal of Medicine and Medical Sciences, Vol. 13 No. 10, 2023, pp. 1580-1583. doi: 10.5923/j.ajmms.20231310.44.
Article Outline
1. Introduction
- In dermatological practice, fungal skin diseases remain relevant, given the continuous rise in their prevalence, particularly among children. Among the causative agents of fungal skin diseases, fungi from the Trichophyton, Microsporum, and Malassezia genera are most commonly observed [1,5]. The high contagion of mycosis agents, the development of infiltrative-purulent forms, chronic courses, and resistance to antifungal therapy necessitate a more profound exploration of the etiopathogenetic mechanisms of mycotic skin infections.A critical aspect in the pathogenesis of fungal diseases is the skin's barrier function. Studies have shown that disruptions in the skin's barrier function in various skin diseases, particularly atopic dermatitis, are attributed to mutations in the gene encoding filaggrin (FLG), part of the epidermal differentiation complex [3,4,7,9-12].Filaggrin is the primary protein responsible for epidermal cell differentiation and the execution of its barrier function. In the stratum corneum, filaggrin molecules, rich in arginine, undergo deimination, converting positively charged arginine residues into neutral citrulline. These molecules disassociate from keratin and degrade into various amino acid components, including pyrrolidone carboxylic acid and urocanic acid [2,3,4].In cases where filaggrin is reduced, as in atopic dermatitis, or absent, as in ichthyosis, the skin's barrier quality deteriorates due to the stratum corneum's inability to retain moisture [4,7,9].This has sparked significant interest in investigating the filaggrin gene (FLG) in developing fungal skin infections in children. The search for genetic factors will help uncover the primary mechanisms underlying the pathogenesis of fungal diseases [6].Objective:The study aimed to explore allelic variants and associations of the 2282del4 polymorphism genotypes in the FLG gene among children with Trichophytosis in the Uzbek population.
2. Materials and Methods
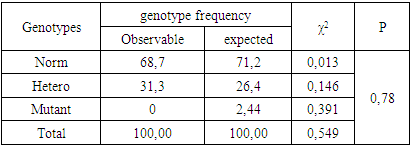
- We examined 16 children aged 1 to 18 years with fungal skin and scalp diseases, with 10 (62.5%) girls and 6 (37.5%) boys. Clinical, immunological, molecular-genetic, and statistical studies were conducted for all children. The diagnosis of skin mycosis was made by ICD-10 (International Classification of Diseases, 10th revision) codes, with the primary code being B35.0 [8].Molecular-genetic biomaterials (DNA) examinations were carried out at the "Geno-technology" LLC. The study involved DNA samples from children with fungal skin diseases, who constituted the primary group, and healthy children without fungal skin diseases (the control group). Genetic research was performed following informed consent from the patients. DNA samples were extracted from peripheral blood lymphocytes using a modified methodology. The concentration and purity of the isolated DNA were assessed by measuring the optical density of DNA-containing solutions at wavelengths of 260 and 280 nm against TE (Tris-EDTA) on a NanoDrop 2000 spectrophotometer (USA).Genotyping of the FLG gene's 2282del4 polymorphism was performed using a real-time PCR amplifier Rotor-Gene 6000 Model 65Н0-100 (Australia) with the test system from "Syntol" Cat. No. - NP_555_100_RG (Russia), following the manufacturer's instructions. Statistical analysis of the results was conducted using the statistical software package "OpenEpi 2009, Version 2.3."Molecular-genetic data, including an assessment of the deviation of the studied DNA polymorphism distributions from the Hardy-Weinberg equilibrium (HWE), were analyzed using the "GenePop" (Genetics of Population) software.Allele and genotype frequencies (f) were calculated using the formula:f = n/2N and f = n/N,where n is the frequency of the variant (allele or genotype), and N is the sample size.Allele frequencies were calculated using the formula:P = (2N|+N2)/2N, q = (2N3+N2)/2N,where P represents the frequency of allele A, q is the frequency of allele a, and N is the total sample size (N=N|+N2+N3), where N|, N2, N3 are the counts of individuals with genotypes AA, Aa, and aa, respectively.To calculate the odds ratio (OR) with a 95% confidence interval (CI), χ2, and p-values, the statistical software package "OpenEpi 2009, Version 2.3" was used.The relative deviation of observed heterozygosity from expected heterozygosity (D) was calculated using the formula:D = (hobs-hexp)/hexp,where hobs and hexp represent the observed and expected heterozygosity, respectively.The prognostic efficiency (AUC - area under the curve) of the studied genetic markers was determined using the standard formula:AUC = (Se + Sp)/2,Where Se and Sp are the sensitivity and specificity of the genetic marker, respectively, if the AUC value is <0.5, the marker is a random classifier; AUC=0.5–0.6 indicates a poor classifier; AUC=0.6–0.7 is a moderate classifier; AUC=0.7–0.8 is a good classifier, and AUC>0.8 represents an excellent classifier.Statistical significance was accepted at p<0.05.
3. Results
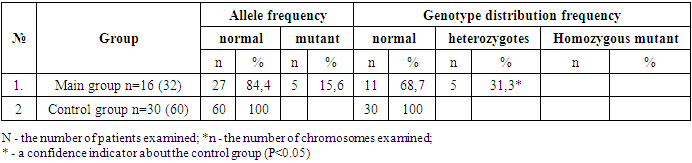
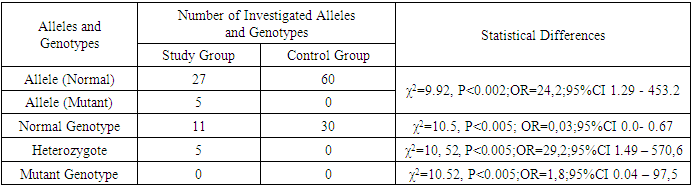
- In terms of clinical forms, among the 16 children, 14 were diagnosed with smooth skin trichophytosis and trichophytosis of the scalp, while 2 had microsporia of the smooth skin. Among the 14 children with trichophytosis, five were diagnosed with the superficial form of smooth skin trichophytosis, and 9 had the infiltrative-purulent form of scalp trichophytosis. All children were included in the combined group for laboratory analysis. The results of the molecular-genetic investigations of the FLG gene's 2282del4 polymorphism are presented in Table 1.As evident from the table, a comparative analysis of allele and genotype frequency distribution of the 2282del4 FLG gene polymorphism among 60 DNA samples from 30 healthy children revealed the presence of the normal gene allele in 100% of cases (60/60, N=60, n=60), whereas in the primary group of children with trichophytosis, among 114 DNA samples (N=114, n=32), the normal allele was identified in 84.4% (27/32), which was 1.2 times lower compared to the control group (*χ2=9.91, P<0.002; OR=0.04; 95% CI 0.0-0.77). Meanwhile, the mutant allele of the 2282del4 FLG gene was detected in 15.6% of cases (5/32) in the primary group of children with trichophytosis, whereas it was not observed in the control group (*χ2=9.92, P<0.002; OR=24.2; 95% CI 1.29 - 453.2).
|
|
|
4. Conclusions
- 1. Molecular-genetic analysis of the frequency distribution of 2282del4 FLG gene alleles in children with fungal diseases revealed the presence of the mutant allele variant in 15.6% of cases (5/32) (χ2=9.92, p<0.002; OR=24.2; 95% CI 1.29 - 453.2).2. Heterozygous variants of the 2282del4 FLG gene polymorphism were detected in 31.3% of cases (5/16) in the study group of children with trichophytosis, whereas none were found in the control group (χ2=10.52, p<0.005; OR=29.2; 95% CI 1.49 – 570.6). Mutant homozygous variants of the 2282del4 FLG gene polymorphism were not identified in either group (χ2=10.52, p<0.005; OR=1.8; 95% CI 0.04 – 97.5).3. Heterozygous genotype variants of the 2282del4 FLG gene polymorphism are a significant marker of increased risk for the development of fungal diseases caused by dermatophytes of the Trichophyton and Microsporum genera in Uzbek children (χ2=10.52, p<0.005; OR=29.2; 95% CI 1.49 – 570.6). Functionally favorable genotype 2282del4 FLG gene variants serve as reliable protective markers against the development of pathology (χ2=10.5, p<0.005; OR=0.03; 95% CI 0.0-0.67).
5. Study Limitations
- Within the scope of this study, no significant limitations that could impact the accuracy and generalizability of the obtained results have been identified.
ACKNOWLEDGEMENTS
- This article does not mention any specific acknowledgments or expressions of gratitude.
Funding
- This research has not received financial support from external sources or organizations.
Conflict of Interest
- The authors of this article confirm the absence of any conflicts of interest that could influence the research findings.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML