-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2023; 13(9): 1213-1217
doi:10.5923/j.ajmms.20231309.07
Received: Aug. 30, 2023; Accepted: Sep. 21, 2023; Published: Sep. 22, 2023

Analysis of the Role of TNF-a Gene Rs1800629 Polymorphism in Disease Development in COVID-19 Infected Pregnant Women
Shahnoza Alimjanovna Zufarova1, Mehrinoz Oybekjon Qizi Komilova2, Qodirjon Tukhtabaevich Boboev3, Abdusalomova Zukhra Khayrullo Qizi4
1Doctor of Medical Sciences, Director of the Republican Population Reproductive Health Center, Tashkent, Uzbekistan
2PhD Student, Andijan State Medical Institute, 2- Department of Obstetrics and Gynecology, Andijan, Uzbekistan
3Doctor of Medical Sciences, Professor Head of the Department of Molecular Medicine and Cell Technologies Republican Specialized Scientific and Practical Medical Center of Hematology, Tashkent, Uzbekistan
4Student, Andijan State Medical Institute, Therapeutical Faculty, Andijan, Uzbekistan
Correspondence to: Mehrinoz Oybekjon Qizi Komilova, PhD Student, Andijan State Medical Institute, 2- Department of Obstetrics and Gynecology, Andijan, Uzbekistan.
| Email: |  |
Copyright © 2023 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

The infection of COVID-19 is a disease that is common worldwide and can cause acute respiratory failure and systemic complications, and the study ofits pathogenesis can prevent complications of the disease and create opportunities for individual work with patients. The purpose of the study: to study the role of tumor necrosis factor TNF-α rs1800629 polymorphism in the pathogenesis of the development of COVID-19. The frequency of detection of TNF-α rs1800629 polymorphism in patients with severe and mild form of COVID-19 was studied in samples of patients and conditionally healthy controls. Real-time PCR analysis was performed on a DNA sample obtained from peripheral blood. Weak genotypic variants of the TNF-α gene rs1800629 polymorphism were compared with the main group of patients and control samples. The results showed that the G/C genotype of TNF-α rs1800629 polymorphism was no significantly higher in women with more severe coronavirus disease compared to healthy donor samples (χ2=0.4; p=0.6; RR =1.2; 95% CI: 0.64 - 2.45; OR=1.3; 95%CI: 0.6 - 2.81) and the G/G genotype was less defined in the patient group compared to the control sample (χ2=0.4; p=0.60; RR=1.0; 95%CI: from 0.49 to 1.89; OR=0.8; 95% CI: 0.36-1.68). Conclusion: Frequencies of TNF-α rs1800629 polymorphism genotypes do not play a role in the pathogenesis of the development of COVID-19 infection.
Keywords: Genotype, TNF-α, Allele, Cytokines, Chemokine, ARDS, Polymorphism
Cite this paper: Shahnoza Alimjanovna Zufarova, Mehrinoz Oybekjon Qizi Komilova, Qodirjon Tukhtabaevich Boboev, Abdusalomova Zukhra Khayrullo Qizi, Analysis of the Role of TNF-a Gene Rs1800629 Polymorphism in Disease Development in COVID-19 Infected Pregnant Women, American Journal of Medicine and Medical Sciences, Vol. 13 No. 9, 2023, pp. 1213-1217. doi: 10.5923/j.ajmms.20231309.07.
Article Outline
1. Introduction
- Currently, many scientists are interested in determining the role of genetic factors in the development of COVID-19 infection. Identification of genetic factors for predicting infectious susceptibility and disease severity is important, as it allows identification of pathophysiological changes in viral infections and potential drug targets [1]. In addition, it can reveal causal relationships between risk factors, biomarkers, and disease, and shed light on prevention knowledge [1]. Recent statistics among all COVID-19 patients show that 19% are severe, and most of them have significantly increased levels of pro-inflammatory cytokines and subsets of immune cells [2,3,4]. Although many vaccines have been developed, uncertainty about vaccine safety remains, and how to effectively treat patients with COVID-19 is still a concern [5].Research in the world does not deny that the acceleration of the inflammatory process, the storm of cytokines and the state of the immune system are related to the disease of COVID-19. TNF-α is produced by a variety of cells, including macrophages, monocytes, neutrophils, T cells, and NK cells. TNF-α is located between the HLA-B and HLA-DR genes in the major histocompatibility complex class III region on chromosome 6 [6]. TNF-α is a key pleiotropic mediator of acute and chronic systemic inflammatory responses, regulating cell apoptosis and proliferation while simultaneously stimulating the production of other chemokines and cytokines. TNF-α signaling is essential for the activation of innate immunity against infectious agents, and disruption of its signaling is associated with the onset of cytokine storm and increased risk of death in patients with COVID-19. Post-mortem reports of lung lesions of etiological COVID-19 have revealed pulmonary edema with bilateral widespread alveolar destruction and hyaline membrane development [7,8]. High levels of the pro-inflammatory cytokine TNF-α in severe COVID-19 are associated with cytokine-driven inflammation leading to tissue destruction and pulmonary edema, endothelial cell breakdown is assumed. [9,10].Research in the world does not deny that the acceleration of the inflammatory process, the storm of cytokines and the state of the immune system are associated with the disease of COVID-19 [10]. In addition to the above, TNF-α is involved in physiological processes such as antitumor responses, control of the inflammatory process, and homeostasis of the immune system. [11,12]. TNF-α is one of the most important pro-inflammatory cytokines of the immune system, and dysregulated TNF-α signaling can trigger a cytokine cascade [13]. Therefore, identification of changes in TNF-α gene may be an important factor in explaining the pathogenesis of COVID-19 exacerbation.In recent studies, the TNF-α gene G-308A polymorphism and the TNF receptor have been shown to be associated with outbreaks of Covid-19 infection in Egyptian and Mexican populations, respectively [14]. This study aims to determine the association of TNF-α gene G-308A polymorphism with severity of viral infection in pregnant women in the Uzbek population infected with Covid-19.
2. The Purpose of the Study
- To study the role of the TNF-a gene rs1800629 polymorphism in the pathogenesis of disease severity in pregnant women infected with COVID-19.
3. Materials and Methods
- 110 women at different stages of pregnancy who were infected with different degrees of coronavirus and were not vaccinated against COVID-19 participated in the study. The average age among these pregnant women is 29.35±0.56 years. Venous blood in the amount of 2-3 ml was taken from them for research.Molecular-genetic analysis was studied in a case-control sample. As a control group, blood samples of 105 conditionally healthy women of Uzbek nationality who were not infected with COVID-19 and had no symptoms of coronavirus were assigned.Tumor necrosis factor (TNF-α) gene polymorphism was studied by SNP method using SPF "Litekh" SPN express test systems.Checking the genotyping of characters consists of several stages:1. Peripheral blood sampling.2. Isolation of DNA molecule from lymphocytes.3. Detection of polymorphisms using PCR;4. Visualization of results.Biomaterial sampling was performed using standard vacuum tubes containing EDTA-K3 anticoagulant (Vacutainer Becton Dickinson International, USA). For PCR studies, genomic DNA was isolated using the AmpliPrime RIBO-prep reagent kit (NextBio, Russia). Genomic DNA concentration was measured in a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, USA) at A260/280 nm wavelength. The purity of all DNA samples was 1.7/1.8.PCR analysis was performed using Rotor Gene Q (Guagen, Germany) and Applied Biosystems (model 2720, USA) instruments and according to the amplification programs.The statistical significance of the data collected as a result of the research was studied using the 2021 program of the R Microsoft office, Epi info software package.In order to assess the relationship between the result and risk factors, the 95% confidence interval for the exploration of the biostatistics risk ratio, in order to determine the statistical significance of the obtained results, X2 and R were calculated according to the Pearson test. 95% confidence intervals were represented in the charts.Statistical processing of clinical material was carried out using the statistical package of "STATISTICA 10.0" applications.A total of 110 pregnant women infected with the coronavirus disease taken for the study formed the main group, this group was divided into 2 subgroups depending on the severity of the disease:1. subgroup A - patients with (severe) COVID-19 complicated by pneumonia (n=70).;2. subgroup B - pregnant women mildly infected with COVID-19 (n=40).
4. Results
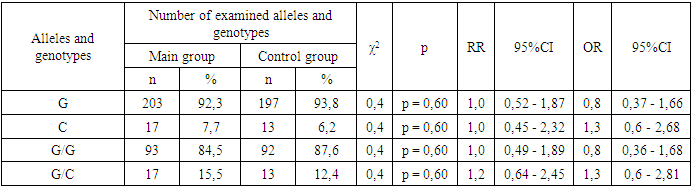
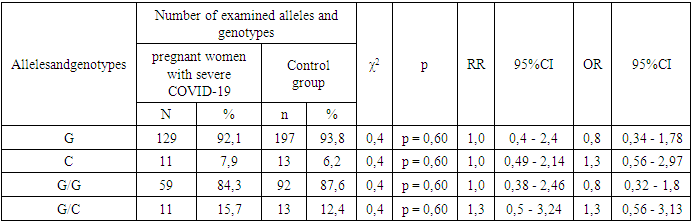
- Frequencies of alleles and genotypes of the TNF-α gene rs1800629 polymorphism in the main group of pregnant women with different manifestations of COVID-19 and the proportions in the control group. According to it, the correlation between G and C alleles of TNF-α gene rs1800629 polymorphism in the main and control was 92.3% to 7.7% and 93.8% to 6.2% . In the control group of women infected with COVID-19, the dominant G/G genotype was 84.5% compared to 87.6% in the control group, and the heterozygous G/C genotype was 15.5% and 12.4%.In the study of the genotypes of the TNF-a gene rs1800629 polymorphism, it was found that in the main group of 110 pregnant women infected with COVID-19, the dominant homozygous G/G genotype was 84.5%, in the control group it was 87.6%. In statistical analysis, it was found that this genotype is insignificant in the origin of COVID-19 (χ2=0.4; p =0.60; RR=1.0; 95%CI: 0.49-1.89; OR=0.8; 95% CI 0.36–1.68). The frequency of detection of the heterozygous G/C genotype was 15.5% and 12.4% in women infected with COVID-19 and in the control group, respectively. According to the odds ratio, in the analyzes of the TNF-α gene rs1800629 polymorphism, the heterozygous G/C genotype had statistically insignificant results in the development of the disease: χ2=0.4; p=0.60; RR=1.2; 95%CI: 0.64-2.45; OR=1.3; 95%CI: 0.6–2.81) (Table 1).
|
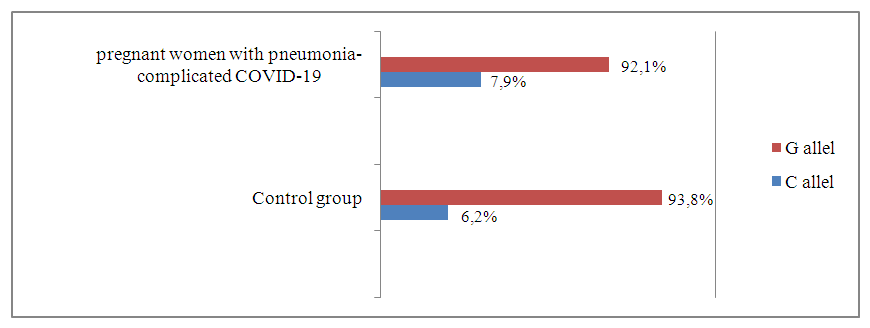 | Figure 1. Prevalence of TNF-α gene rs 1800629 polymorphism alleles in pregnant women infected with COVID-19 complicated by pneumonia and control groups |
|
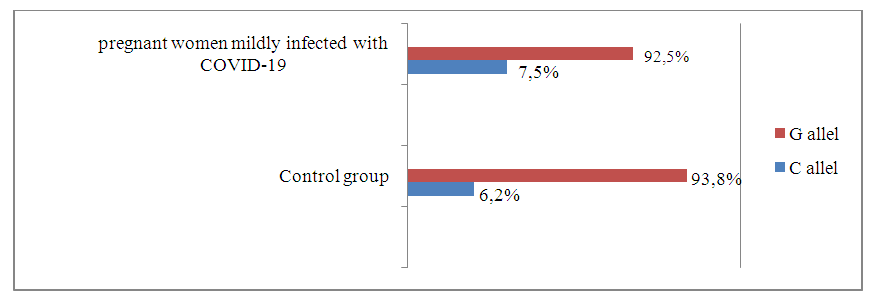 | Figure 2. Prevalence of TNF-α gene rs1800629 polymorphism alleles in pregnant women mildly infected with COVID-19 and control groups |
|
5. Discussion
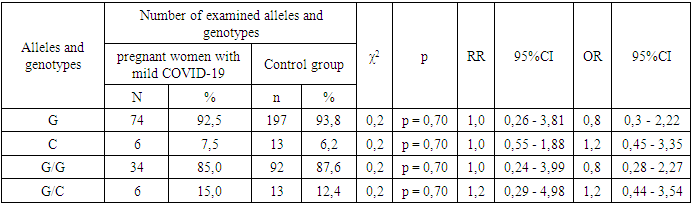
- According to the results of the study, there are no significant differences in the distribution of alleles and polymorphisms of the TNF-α gene rs1800629 polymorphism between the main group of pregnant women infected with COVID-19 and the samples of the healthy group. The minor allele C was 1.3 times more frequent in the baseline group than in the control group and had a tendency to increase the risk of developing the disease by 1.3 times (7.7% and 6.2%, respectively; χ2=0.4; P=0.60; RR=1.0; 95%CI: 0.45–2.32; OR=1.3; 95%CI: 0.6–2.68). In the analysis of the prevalence of the weak G/G genotype of the TNF-α gene, comparing healthy donor samples with the main group samples, it was 87.6% versus 84.5%, respectively, and statistical analyzes showed that the homozygous G/G genotype had not a protective effect against the disease (χ2=0.4; p=0.60; RR=1.0; 95%CI: 0.49-1.89; OR=0.8; 95%CI: 0.36-1.68). There was a slight difference in the determination of the heterozygous G/C genotype in patients and controls: 15.5% and 12.4%. Statistical analyze showed that this genotype was only 1.2 times more frequent in patients with COVID-19 and statistically insignificant (χ2=0.4; p=0.60; RR=1.2; 95%CI: 0.64–2.45; OR=1.3; 95%CI: 0.6–2.81). Also, comparing the samples of women with pneumonia and those of the control group, no statistically significant results were obtained (p>0.05), which indicates that TNF-α gene rs1800629 polymorphism does not play a role in the development of pneumonia in women infected with COVID-19. The mutant C/C genotype of this polymorphism was not found in the groups due to its lethal nature.When analyzing the literature, there are many studies devoted to determining the role of TNF-α gene rs1800629 polymorphism in the development and complications of COVID-19, but the association of this gene with pregnant women and COVID-19 infection has not been investigated. The results of our study are consistent with the data of Wang S., Wei M. et al [15]. In his research, they found that polymorphisms of the TNF-a gene do not play a role in the development of the coronavirus disease. But in the studies of Ahmad Salih and a number of other scientists, the C allele is listed as a risk factor for the development of the disease [16,17]. Our study did not agree with these findings. However, large-scale regional and individual studies are required to draw conclusions about the role of cytokine-responsive gene polymorphisms in the severity of COVID-19.
6. Summary
- The role of alleles and genotypes of the TNF-α gene rs1800629 polymorphism in the development of COVID-19 and the origin of pneumonia was studied and analyzed in pregnant women. When the distribution of alleles was studied, it was found that the dominant G allele does not have the property of protecting against the disease, and the minor C allele is insignificant in the development of the disease. According to the results of molecular genetic examination and statistical research, we can conclude that the heterozygous G/C genotype of the TNF-a gene does not play a role in the pathogenesis of mild and severe forms of COVID-19. No statistically significant results were obtained when the main group and subgroups were compared to the control sample (p>0.05). The homozygous G/G genotype did not have a protective effect against the development of severe and mild forms of COVID-19 and the development of pneumonia.The final conclusions proved that it is impossible to use the TNF-α gene rs1800629 polymorphism as a reliable marker in predicting the exacerbation of the coronavirus disease and complications with pneumonia.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML