-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2023; 13(5): 626-632
doi:10.5923/j.ajmms.20231305.17
Received: Apr. 20, 2023; Accepted: May 9, 2023; Published: May 17, 2023

Prevalence of eNOS 3 Gene Polymorphism in Family Members of Patients with Familial Bronchial Asthma in the Uzbek Population
Oqboev T. A., Safarova M. P.
Department of Internal Medicine, Samarkand State Medical University, Uzbekistan
Copyright © 2023 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

The frequency of distribution of alleles and genotypes of the polymorphic variant of the eNOS3 gene has been studied in patients with bronchial asthma and healthy individuals in a family of Uzbek ethnicity. In this, 193 individuals in the family were under surveillance. Patient testing was carried out according to the International Classification of the World Health Organization (ICD-10) and the relevant diagnostic criteria of the global strategy for treatment and Prevention of BA (GINA 2021). A polymorphic variant of the eNOS3 gene was investigated by the polymerase chain reaction (PCR) method at the center of “Medical Genetics” of the Institute of Biochemistry of the Academy of Sciences of the Republic of Uzbekistan. In the Uzbek population, patients with family asthma were more commonly encountered in the family carrying the 4b/4b genotype and 4b allele of the eNOS3 gene, and they were more common in allergic types of the disease, mild disease, Disease Control, and women. Homozygous genotype carriers of 4a/4a in the family have been noted to be a non-allergic type of family BA, severe course, and abundant in those with uncontrolled disease. This suggests that the eNOS3 gene in the Uzbek population has a higher risk of developing the disease in these genotypes and allele carriers, which makes it possible to identify the disease early in the family, make a comparative diagnosis and create new methods of treatment.
Keywords: Familial bronchial asthma, eNOS3 gene, 4b/4b, 4b/4a, 4a/4a genotype
Cite this paper: Oqboev T. A., Safarova M. P., Prevalence of eNOS 3 Gene Polymorphism in Family Members of Patients with Familial Bronchial Asthma in the Uzbek Population, American Journal of Medicine and Medical Sciences, Vol. 13 No. 5, 2023, pp. 626-632. doi: 10.5923/j.ajmms.20231305.17.
Article Outline
1. Introduction
- Family – specific genetico-epidemiological studies have found a high incidence of bronchial asthma (BA) among family – members. If one parent has bronchial asthma the risk of developing the disease in their children is three times higher than in a healthy family, if both parents have bronchial asthma it has been found that their children have a six times higher risk of getting it [Didkowski N.A., Jarów M.A. 2005., Bochkov N. P. 2018]. Literature records that 46.3% of individuals in the family were diagnosed with bronchial asthma [Asanov A.Yu.2018]. In Russia I. A study by Cherkashinai et al showed that 18% of family members were diagnosed with BA disease. Ubaydullaev A. M, Yakimova M. A. in a genetic study conducted by a family of patients with bronchial asthma in the Uzbek population for the first time, they studied the prevalence and hereditary predisposition of the disease in Uzbek families. In this, inbred found that in a family with a marriage, bronchial asthma is severe, and the disease occurs early. The distribution of eNOS3 gene genotypes and alleles in different ethnic populations in the world has been studied. In the Turkish population, 4b/4b - 69.0%, 4b/4a- 28.2%, 4a/4a- 2.8% were found [Senkal N.2023]. In Europe, whites are more likely to have a b allele, and their citation is respectively: 4b/4b - 0.41; 4b/4a- 0.46; 4a/4a- 0.13. Among people living in the northwest of Russia, 4b/4b - 63.7%; ba4b/4a- 34.5%; 4a/4a-1.8% were found, while Novosibirsk was found to be represented by 4b/4b - 67.9%; 4b/4a-28.9%; 4a/4a-3.3% distribution [Pozdnyakov N.O. 2015].eNOS3 and other genes calculated from candidate genes among individuals in the family – families of patients with hereditary predisposition to bronchial asthma in different ethnic groups are in the cluster of distribution of genotypes and alleles [Kelembet n. A. 2005., Sardaryan I. S. 2009., Cherkashina I.I. 2010., Pasieshvili T. M.2012., Bezrukov L.A. 2015., Shakhanov A. V. 2017., Cortez e Castro M. 2020., Dai X., Inna K. 2021., Agache I, Aline E., Aytac H. M. 2022.] a number of scientific studies have been conducted.In particular, in Saint Petersburg the N.A. Kelembet eNOS3 gene is abundant in the 4a allele and the 4a/4a genotype in the BA medium-heavy species, known as in Astrakhan. X. A. Akhminayev., L.P. Varoninain a study conducted by and co-authors, BA reported a high incidence of 4a/4b polymorphism in patients. Of the 150 patients with BA who spent in Tomsky, 4b/4b - 65.33-63.7%; 4b/4a-32.67-34.5%; 4a/4a-2.00-1.8% were observed [Babushkina N.P. 2015].In patients with bronchial asthma in Uzbekistan, the eNOS3 gene has not been studied before studies on the distribution of genotypes and alleles.
2. Research Objective
- Family eNOS3 gene studies the occurrence of bronchial asthma in genotype and allele-carrying individuals.
3. Materials and Methods
- Accordingly, we studied the distribution of allele and genotype frequencies of the polymorphic variant of the eNOS3 gene in patients with bronchial asthma and healthy individuals of Uzbek nation. 193 individuals were examined in the family. All patients with family BA underwent extensive clinical, functional and laboratory tests. Patients were examined according to the WHO International Classification (ICD-10) and the diagnostic criteria of the global strategy for treatment and Prevention of BA (GINA, 2021).At the “center of Medical Genetics” of the Institute of Biochemistry of the Academy of Sciences of the Republic of Uzbekistan (director, Doctor of medical sciences, professor Mukhamedov R.S.) analysis of the polymorphic variant of the eNOS3 gene was done through the PCR method.The control group is made up of 45 (of which 23 are male and 22 are female) practically healthy people aged 17-62 (average age 28.64) living in the Republic of Uzbekistan.Polymorphisms of genes under study in asthma patients and the control group coincided with The Hardy-Weinberg equilibrium, where the distribution of genotypes was expected. The difference in allele and genotype frequency distribution was evaluated through the χ2 criterion. To assess the Association of alleles and genotypes with the results, the relative risk (OR) was calculated with a 95% confidence interval (CI). Online calculator for statistical processing of molecular genetic verification data (https://medstatistic.ru/index.php).
4. Conclusions and Discussion
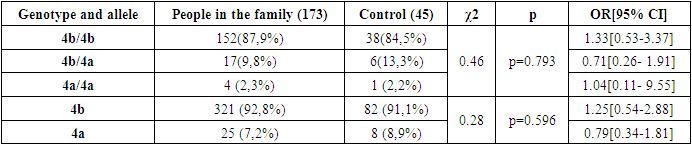
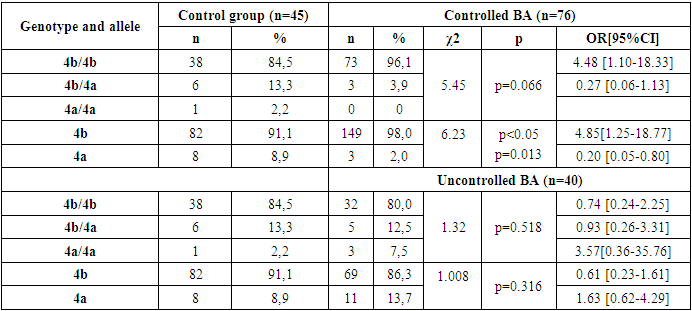
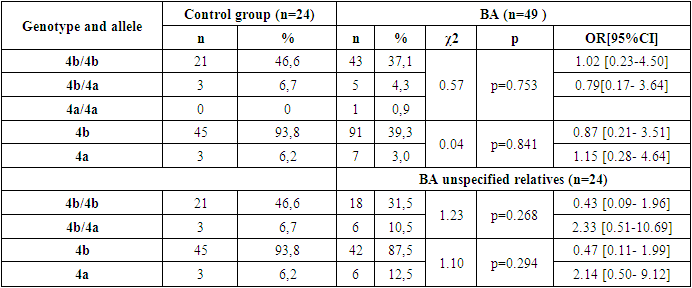
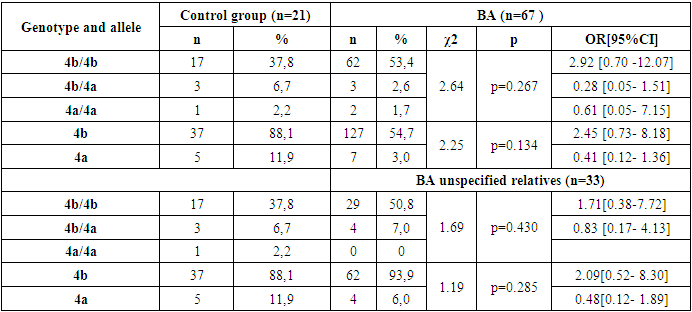
- Based on the results obtained, when the frequency of occurrence of eNOS3 gene genotypes and alleles in 49 family individuals was assessed, the following distribution in 173 individuals in the family was 4b/4b genotype - 152(87.9%), 4b/4a genotype - 17(9.8%), 4a/4a genotype - 4 (2.3%), 4b allele - 321 (92.8%), 4a allele - 25 (7.2%). In 45 practically healthy individuals examined as a control group, however, 4b/4b genotype - 38(84.5%), 4b/4a genotype - 6(13.3%), 4a/4a genotype - 1 (2.2%), 4b allele - 82 (91.1%), 4a allele - 8 (8.9%) were reported [Table 1].
|
|
|
|
|
|
|
5. Conclusions
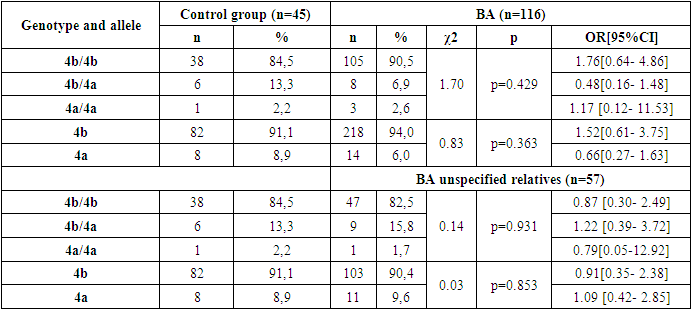
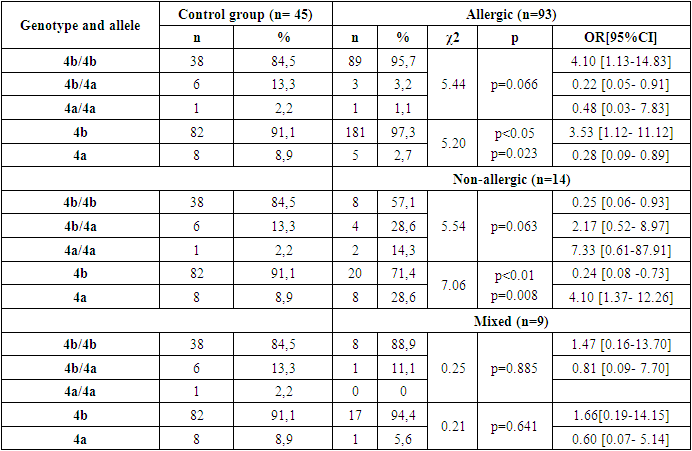
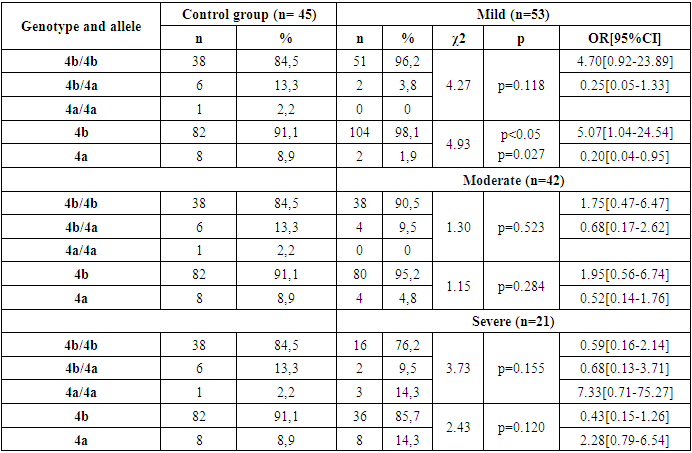
- Thus, in the family, eNOS3 gene 4b/4b genotype and 4b allele carriers were more common for those with familial BA and were more likely to diagnose the disease by allergic type, mild degree of disease, in those with Disease Control, and in women. In homozygous genotype carriers with 4a/4a alleles in the family, those with familial BA were poorly identified and the disease was reported by a not allergic type, those with severe illness, and those without control. This can lead to early detection of the disease in the family, comparative diagnosis and the creation of new treatments, indicating that the eNOS3 gene in the Uzbek population is high in disease progression in carriers of this genotype and allele.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML