-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2023; 13(1): 18-23
doi:10.5923/j.ajmms.20231301.05
Received: Dec. 14, 2022; Accepted: Dec. 29, 2022; Published: Jan. 13, 2023

Distribution of Bacterial and Fungal Coinfection in COVID-19
L. Tuychiev1, J. Tuychiev1, 2, G. Abdukhalilova2
1Department of Infectious and Pediatric Infectious Diseases, Tashkent Medical Academy, Tashkent, Uzbekistan
2Republican Specialized Scientific Practical Medical Center of Epidemiology, Microbiology, Infectious and Parasitic Diseases, Tashkent, Uzbekistan
Correspondence to: L. Tuychiev, Department of Infectious and Pediatric Infectious Diseases, Tashkent Medical Academy, Tashkent, Uzbekistan.
| Email: |  |
Copyright © 2023 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Microbial coinfection raises the chance of illness severity in people with new coronavirus infection, according to worldwide statistics. A study was conducted to determine the prevalence of bacterial and fungal coinfection in hospitalized patients with confirmed SARS Coronavirus 2 infection (SARS-CoV-2). The results of a microbiological examination of 672 laboratory-confirmed COVID-19 patients who underwent inpatient treatment at the Republican Specialized Scientific-Practical Medical Center of Epidemiology, Microbiology, Infectious and Parasitic Disease (RSSPMCEMIPD) and specialized coronavirus hospital “Zangiata #1” in Tashkent in 2020, 2021 and 2022, were encompassed in this cross-sectional study. Eight pathogens were found in patients, and 232 (34.5%) of them were infected with one or more of them. Klebsiella pneumoniae was the most prevalent pathogenic bacteria, followed by Staphylococcus aureus. There were no correlations between the occurrence of coinfection and age groups, gender, or illness severity. These results will be useful reference materials for the diagnosis and clinical management of COVID-19 patients.
Keywords: COVID-19, Coinfection, Severity, Mortality
Cite this paper: L. Tuychiev, J. Tuychiev, G. Abdukhalilova, Distribution of Bacterial and Fungal Coinfection in COVID-19, American Journal of Medicine and Medical Sciences, Vol. 13 No. 1, 2023, pp. 18-23. doi: 10.5923/j.ajmms.20231301.05.
Article Outline
1. Introduction
- In Wuhan, Hubei Province, China, in 2019, a novel coronavirus (SARS-CoV-2) was reported, which has never been encountered in humans preliminary [1]. The World Health Organization labeled this epidemic a public health emergency of worldwide concern on January 30, 2020, and a pandemic on March 11 [2]. Over 650 million instances of the illness had been reported globally as of December 1, 2022; more than 6.6 million individuals had died, and more than 625 million had recovered [3]. The present COVID-19 infection outbreaks serve as a reminder that coronaviruses are a severe and life-threatening menace to worldwide public health. To date, there is no proven antiviral treatment, although over two years have passed since the first case of infection was detected [4]. The above circumstance highlights the control of the infection source, early diagnosis, quarantine measures, supportive treatment and the vaccination as the best way to combat the dire and critical SARS-CoV-2 infections [5]. Since pathogens present similar clinical signs, it is problematic for medical practitioners to differentiate pathogens without a laboratory diagnosis due to the range of airborne illnesses. However, a large number of research focused solely on SARS-CoV-2, ignoring the fact that coinfection by specific viruses might preclude proper illness identification. Yet, the kinds of co-infected pathogens and the proportion of coinfection in SARS-CoV-2 patients are incompletely understood. In the period 2020 - 2022 we investigated the microbiological parameters of coinfection in 672 patients who underwent inpatient therapy at the RSSPMCEMIPD and specialized coronavirus hospital “Zangiata #1” in Tashkent. This study will serve as a blueprint for organizing coinfection prevention, patient care, and empirical therapy in Uzbekistan. The objective of the present study: to determine the distribution of bacterial and fungal coinfection in COVID-19 patients.
2. Material and Methods
2.1. Clinical Data and Specimens Collection
- In the period 2020 - 2022, 672 sputum samples were obtained at random from patients with a confirmed COVID-19 diagnosis and receiving inpatient therapy at the RSSPMCEMIPD. Samples were collected and promptly submitted for seeding to the bacteriological laboratory at the ISO 15189-accredited Center of Antimicrobial Resistance. Medical records were used to acquire clinical, laboratory, and baseline data. At the time of admission, the following characteristics were recorded: age, gender, illness severity, clinical symptoms, and results of laboratory and instrumental examinations. The Ethics Committees of the Ministry of Health of Uzbekistan and the RSSPMCEMIPD authorized all methods used in this investigation that encompassed human materials.
2.2. Identification of Bacterial and Fungal Infections
- SARS-CoV-2 was confirmed by real-time RT-PCR. Secondary bacterial and fungal infections were identified by sowing sputum on special nutrient media:ü Blood agar with a disk with optochin in the center of the nutrient medium.ü Chocolate agar (or selective blood agar for H. influenza).ü Bile-esculin agar ü Milk-salt agarü Agar McConkeyü Agar SaburoDishes with cultures on blood and chocolate agar were incubated in an atmosphere CO2 5% at 35-37 °C 48h; dishes with cultures on McConkey agar were incubated under normal circumstances. Gram-positive cocci were cultivated on mannitol-salt agar and agar with 5% sheep blood. Additionally, bile-esculin agar was used to differentiate enterococci. In the presence of gram-positive yeast-like structures in the stained smear, tubes with Saburo agar with dextrose were used for cultivating the samples. The identification of microorganisms was carried out by the classical method using the sets EnteroPlu test, Staf system 18R, Streprosystem 12R, manufactured by Liofilchem, Italy.
2.3. Statistical Analysis
- The Kruskal-Wallis H-test was used to compare continuous variables between groups, and the Dunnett test was employed for paired comparisons; categorical variables were represented as a number (percent) using the Chi-square criteria or the precise Fisher criterion. The significance threshold was set at P 0.05. SPSS 26.0 software was used for statistical analysis.
3. Results
3.1. Patient Characteristics
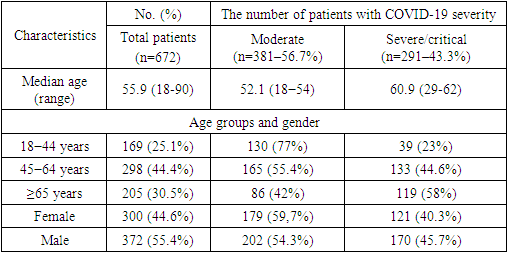
- Table 1 lists the characteristics of all cases and distinct groups. All 672 patients were diagnosed with SARS-CoV-2 infection after PCR confirmation, and their clinical severity was determined using the revised diagnostic criteria of the Ministry of Health of the Republic of Uzbekistan's Interim National Protocols for Diagnosis and Management Patients with a New Type of Coronavirus Infection.
|
3.2. Coinfection of SARS-CoV-2 with Secondary Pathogens
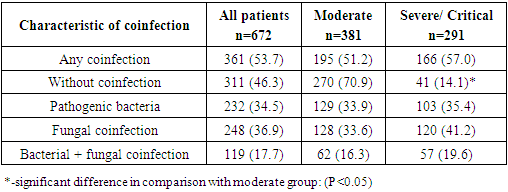
- Out of 672 COVID-19 patients, 361 (53.7%) had simultaneous bacterial and fungal infection, according to our research. Pathogenic bacterial infections were found in 232 (34.5%) patients, whereas fungal pathogens (C. albicans) were found in 248 (36.9%), and mixed infections (bacteria + fungus) were found in 119 (17.7%) patients (Table 2).
|
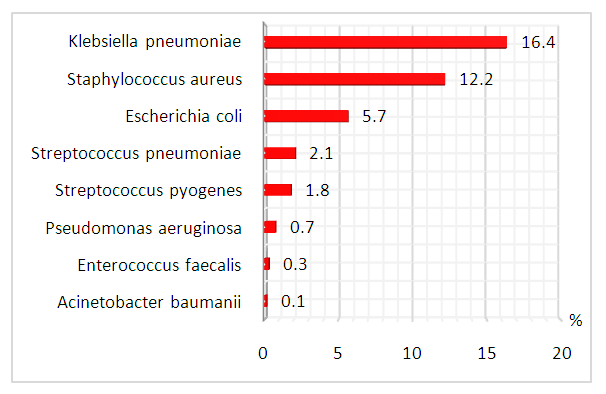 | Figure 1. Distribution of pathogenic bacteria in COVID-19 patients (%) |
3.3. Coinfection Pattern Concerning Different Clinical Types
- Among all isolated pathogenic bacteria, the proportion of K.pneumoniae was the highest and the same in the group of patients with severe/critical and moderate COVID-19 - 16%. S.aureus and E.coli were determined more often in moderate cases (13.9%) than in the group with severe/critical disease (10%). Three species of pathogens (Str.pneumoniae, Str.pyogenes, P.aeruginosa) were more often detected in patients with severe illness (P>0.05), while E.faecalis and A.baumanii were detected only in severe patients. C.albicans was diagnosed more often in patients with severe course of disease - 120 (41.2%) and relatively less in moderate COVID-19 (33.6%).
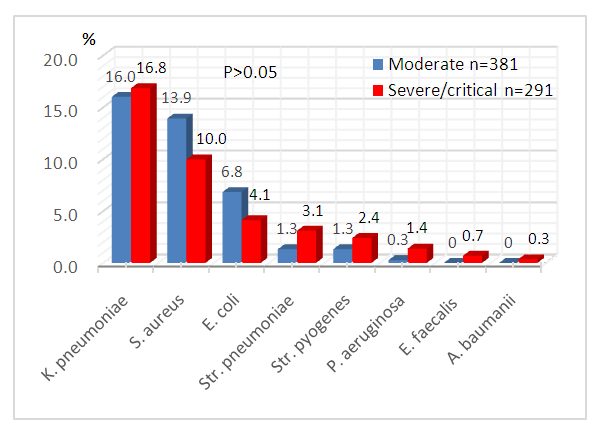 | Figure 2. Bacterial coinfection of patients with various severity of COVID-19 (%) |
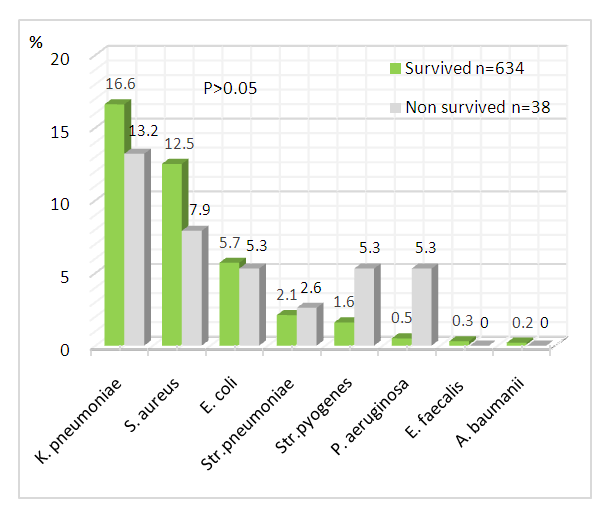 | Figure 3. Bacterial coinfection depending on outcome of patients with COVID-19 (%) |
4. Discussion
- In individuals with coronavirus infection, the current medical literature finds modest incidence of bacterial and fungal coinfection. In their meta-analysis, Rawson and colleagues identified an 8% incidence of bacterial and fungal coinfection in patients with coronavirus infection, whereas the usage of empirical antibacterial treatment was 72% [7]. Today, it is conspicuous that subsequent coinfection is uncommon, and that widespread usage of antibacterial medications may only inhibit patients' health. The selection and administration of antimicrobial therapy for respiratory bacterial - fungal coinfection come to the forefront in the treatment of individuals infected with a novel form of coronavirus. Concerns were also expressed about the contingency of abrupt cardiac arrest caused by the lengthening of the QT interval, which is linked with several of the medications used to treat coinfection [6].Another issue connected with the rapid growth of SARS - COV-2 treatment facilities is the possible rise in the incidence of nosocomial infection [7]. Unwittingly, we were not able to test nosocomial infectious agents. Along with nosocomial illness, there have been several instances of patients on mechanical lung ventilation becoming infected [8,9,12]. The techniques for collecting bronchoalveolar lavage fluids were also unavailable for our investigation in order to avoid the creation of aerosols. Notwithstanding this, the guidelines should focus on maintaining adequate infection control of antimicrobial usage and effective antimicrobial resistance surveillance at this time, when the SARS-COV-2 pandemic is spreading and putting a great strain on health systems. Taking actions to reduce antibiotic usage is becoming increasingly obvious during the COVID-19 pandemic [7]. Since the pandemic has induced a significant shortage of pharmaceuticals, particularly crucial antimicrobials, cautious antimicrobial usage will be critical to ensuring that those with established bacterial illnesses receive treatment [17,18]. The safety of early oral antibiotics compared to intravenous ones was revealed for several infections, encompassing lower respiratory tract infections [13-16]. Owing to the need for optimal provision of beds for patients with COVID-19, special attention should be paid to the development of guidelines on optimal pharmacokinetic and pharmacodynamic strategies for common infections requiring the use of antimicrobials to support early switching to oral antibiotics and de-escalation of treatment in patients with short-and long-term infections [19,20]. At the same time, it's pivotal to remember that simultaneous bacterial coinfection is the major cause of death in viral pneumonia [21]. It is anticipated that the corollary of our study will help to establish a reasonable strategy to the selection and usage of antimicrobial medicines for COVID-19 coinfection. In patients with COVID-19, a low prevalence of laboratory-confirmed pathogenic bacteria coinfection was found. The majority of hospitalized patients had sputum culture findings available, however, only a few clinically significant infections were identified. According to the statistics, 34.5% of all enrolled patients were afflicted with 8 types of pathogenic bacterial infections and one type of fungal infection. As a result of various complications of COVID-19, 38 patients died. Eight bacteria species characterize the pathogenic microflora, with no evident preference for age, gender, malady severity, or mortality. According to the results of a meta-analysis of 30 studies from the Embase, Medline, Cochrane Library, LILACS, and CINAHL databases, among 3834 patients, the incidence of bacterial infections with COVID-19 was only 7%, and the total proportion with a secondary viral infection was 3% [22].The most prevalent pathogenic bacteria coinfection in all COVID-19 patients, as per the studies, was bacterial coinfection with K.pneumoniae and S.aureus. While one of the infections is opportunistic, cases of co-colonization have also been documented. The review data on identification of K.pneumoniae are from different continents, i.e., Europe, Asia, North and South America and Africa. The prevalence of coinfection in COVID-19 patients ranged from 0.35% to 53%. [23]. In Wuhan, China, Li et al. demonstrated that among 159 strains of bacteria isolated form 102 hospitalised COVID-19 patients with acquired secondary bacterial infections, Acinetobacter baumannii was the most common pathogen (35.8%), followed by Klebsiella pneumoniae (30.8%) [24]. An unexpected finding was that Enterococcus faecalis was detected in the sputum of two patients with a new coronavirus infection. In the literature, there are reports of an increase in the number of enterococci in patients with COVID-19. In one case, a patient with a fever, dyspnea, and cough was diagnosed with nosocomial pneumonia caused by Enterococcus faecalis and tested positive for COVID-19 [25]. Although the precise mechanism remains unknown, the viral infection seems to cause changes in the bacterial microbiome, favoring Enterococcus and decreasing the intestinal barrier, which provides the necessary condition to develop invasive infections [26].There were a few flaws in the study. The findings of our study are confined to the patients' short-term follow-up. Many biases and data gathering are made harder by the observational design. Several patients were unable to collect sputum while hospitalized, and invasive respiratory sampling was not accessible to reduce aerosol production processes. The significance of Candida spp. from respiratory tract samples as being representative of oropharyngeal candidiasis is ambiguous; most patients will have been receiving broad-spectrum antibacterial therapy at the time of culture. Additionally, we hypothesize that patients may have been using antimicrobial drugs prior to admission to the hospital, which could eliminate or weaken secondary bacterial coinfection, or, conversely, contribute to the appearance of fungal flora. The identification of microorganisms was carried out by the traditional microbial method. We couldn’t use 16S rRNA gene sequence analysis, because of technical and cost considerations.
5. Conclusions
- Despite the relatively high rate of microbiologically substantiated microbial coinfection in confirmed SARS-CoV-2 patients in our investigation, no significant association was reported between coinfection and severity, mortality, or other indicators. This reaffirms that antibiotic therapy should only be prescribed when indicated, following local guidelines, and verified with a clinical response within 48–72 hours. Antibiotic medication is to be stopped if no indication of bacterial coinfection is identified. Late secondary bacterial and fungal infections are less prevalent, and more study is needed to determine their occurrence, nature, and impact.
ACKNOWLEDGEMENTS
- The authors appreciate the role played by the Center of Antimicrobial Resistance of Republican Specialized Scientific Practical Medical Center of Epidemiology, Microbiology, Infectious and Parasitic Diseases, Tashkent.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML