-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2021; 11(10): 683-689
doi:10.5923/j.ajmms.20211110.05
Received: Sep. 13, 2021; Accepted: Oct. 12, 2021; Published: Oct. 30, 2021

The Relationship between the Polymorphism of the MDR1 and MTHFR Genes and the Development of Rheumatoid Arthritis in the Uzbek Population
Ziyadullaev Shukhrat Khudaiberdievich1, Sultanov Ilkhom Islamovich1, Nikitina Natalya Mikhaylovna2, Dushanova Gavkhar Abdukarimovna3, Aripova Tamara Uktamovna4, Kamalov Zaynitdin Sayfutdinovich4
1Samarkand State Medical Institute, Uzbekistan
2Samara State Medical University, Samara, Russia
3Samarkand State University, Uzbekistan
4Institute of Immunology and Human Genetic of Academy of Science of the Republic of Uzbekistan
Copyright © 2021 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

This article presents a study of the analysis of the association of MDR1 and MTHFR gene polymorphism with the development of rheumatoid arthritis in individuals of Uzbek ethnicity. The revealed pronounced association of rheumatoid arthritis with the C3435T polymorphic locus of the MDR1 gene for the C677T polymorphic marker of the MTHFR gene proves the absence of an association with rheumatoid arthritis in the studied population.
Keywords: Rheumatoid arthritis, Rheumatoid factor, Immunoinflammation, Citrulline protein, Oligonucleotides, Polymorphism
Cite this paper: Ziyadullaev Shukhrat Khudaiberdievich, Sultanov Ilkhom Islamovich, Nikitina Natalya Mikhaylovna, Dushanova Gavkhar Abdukarimovna, Aripova Tamara Uktamovna, Kamalov Zaynitdin Sayfutdinovich, The Relationship between the Polymorphism of the MDR1 and MTHFR Genes and the Development of Rheumatoid Arthritis in the Uzbek Population, American Journal of Medicine and Medical Sciences, Vol. 11 No. 10, 2021, pp. 683-689. doi: 10.5923/j.ajmms.20211110.05.
1. Introduction
- One of the actual problems of modern rheumatology and clinical immunology is rheumatoid arthritis (RA). Rheumatoid arthritis is an immune-inflammatory process that causes chronic synovial inflammation, which ultimately leads to joint destruction, as well as systemic complications [3]. The prevalence of RA is 0.5-1.0% of the population. From 70 to 80% of patients with RA have autoantibodies, such as rheumatoid factor and antibodies to citrullinated protein, which fully suggests the development of immuno-inflammatory processes [8,11]. Patients with RA have a higher level of comorbidity and mortality, experience chronic pain, and have greater emotional and social problems affecting their quality of life. They also suffer significant financial losses due to high temporary and persistent disability, and at the same time, 27% of patients have signs of disability that manifest themselves during the first 3 years.Genetic studies of RA were initiated approximately 40 years ago, and the HLA gene and many non-HLA risk loci were identified during recent genome-wide association studies (GWAS). Studies on twins have shown that RA has a heredity of about 50-60% outside of ethnicity [4]. For the present time, more than 100 non-HLA loci associated with RA have been identified, but genes such as TNF-a, known as targets for the treatment of RA, have not been identified in GWAS [9].The main human histocompatibility complex or HLA system is a complex of genes that by themselves, as well as through the products encoded by them, perform the most important biological functions, and primarily provide genetic control of the immune response and the interaction of various cellular elements that implement the immune response. Today, more than 30 loci have been identified, which explain about 20% of the variability in the risk of the disease. The vast majority of RA risk alleles have been identified and confirmed in patients of European origin. The most relevant single nucleotide polymorphisms (SNPs) of non-HLA genes associated with PA include PTPN22, IL23R, TRAF1, CTLA4, IRF5, STAT4, CCR6, PADI4 [6].Genetic, hormonal, environmental, immunological, infectious and other factors take an active part in the development of RA. Unlike classical genetic diseases, in which many different genes and their combinations predispose to the development of the disease, RA is a genetically heterogeneous disease, primarily due to genetic imperfection of immunoregulatory processes. Currently, the roles of other genetic factors that are not directly related to HLA-DR are being actively discussed. These include polymorphism of the genes of the enzymes of the folate cycle MTHFR, MTR and MTRR. These genes are the least studied in terms of predisposition to RA. RA-associated loci and disease have yet to be determined [1,5,12,13].Genetic studies of multifactorial diseases (MFD) are largely facilitated by a detailed description of the pathogenesis of the studied diseases. Knowing the molecular mechanisms of disease formation, it can be assumed that those genes whose protein products are most important in this will be "candidates" for the role of susceptibility genes to the disease. The study of their variability in connection with pathological conditions will reveal hereditary molecular features predisposing to the development of the disease.In this regard, the study of the genetic basis of immuno-inflammatory diseases is still an actual objective, since it is still difficult to imagine the whole picture of the interaction of hereditary and environmental factors in the implementation of such a complex pathological phenotype.
2. Materials and Methods
- This study is based on the results of observation of patients with RA conducted in the rheumatology departments of the clinic No. 1 of SamGosMI and the Tashkent City Clinical Hospital No. 3 during the period of inpatient treatment for the period from 2019-2021. 117 patients with RA were included, the diagnosis was established on the basis of anamnesis, physical and clinical laboratory studies according to the criteria of the American Rheumatology Association (ARA) in 1987 and the working classification proposed by the Association of Rheumatologists of Russia (2003).The age of the patients ranged from 30 to 75 years (average age 56.14±11.69 years), with the duration of the disease from 3 months to 30 years (average duration 9.85±3.44 years). Rheumatoid factor (RF) was detected in 89 patients, and the remaining 20 patients were seronegative. Blood was taken from the cubital vein in a horizontal position, in the morning, on an empty stomach (at least 9 hours). To obtain plasma, blood (3 ml) was collected in a single-use test tube (BD Vacutainer, Plymouth, UK) containing an anticoagulant (tri-substituted sodium citrate), immediately after receiving the sample, it was centrifuged for 15 minutes (3000-4000 ob per minute, 4C), plasma was obtained, aliquoted into labeled secondary plastic Eppendorf tubes and placed in a freezer – 20°C.The material for DNA isolation was venous blood from the ulnar vein with a volume of 3-5 ml (Beckton-Dickinson vacuum cleaners were used for blood collection) with an anticoagulant/preservative 15% tricalium EDTA (Ethilendianin-tetraacetic acid). The blood for further processing could be stored for up to 24 hours at a temperature not higher than +4°C.To obtain genomic DNA, a two-stage method of lysis of blood cells was used. By double centrifugation of the entire volume of whole blood in the RC LB buffer (Red cell lysis buffer) at a speed of 1500 rpm, red blood cells were lyzed for 15-20 minutes. The use of RC LB causes osmotic shock of red blood cells, leading to their swelling and further destruction.The supernatant containing the destroyed red blood cells was carefully drained from the test tube; the remainder of the suprapubic part was sucked off. The remaining clot of the leukocyte mixture at the bottom was lysed in the leukocyte lysis buffer TLB (white cells lysis buffer) in an amount depending on the volume of the leukocyte mixture. Prescriptions of leasing buffers:
 Further purification of leukocyte mass lysates is based on the method of alcohol-salt treatment according to S. Miller et al. ((1988) in a modification proposed by the Stanford University Laboratory. To 400 ml of leukocyte mass lysate, 150 ml of 5M NaCl is added, mixed on a shaker and placed in ice for 10-20 minutes, then centrifuged at 1200 rpm for 15 minutes. The supernatant is taken into another Eppendorf-type test tube and 100% ice ethanol is added. With careful shaking, a quaternary chain of the DNA molecule appears in the mixture; the mixture is centrifuged at 1200 rpm for 15 minutes; the supernatant is removed, and the whitish spot remaining at the bottom of the test tube is washed again in 80% ethanol at 1200 rpm for 10 minutes. The supernatant is drained, the alcohol residues are carefully removed, and the test tube is left open until the alcohol completely evaporates (for 12 hours at room temperature or in a thermostat at a temperature of 40-45°C for 2 hours). After the alcohol has evaporated, a TE (Tris-EDTA) solution diluted with distilled water is added to the test tube with dried DNA in a ratio of 1:3 (TE: water) pH 8.0. The DNA was stored at – 20°C.Methods of identification of allelic variations of polymorphic loci of patients' genesPolymerase chain reaction (PCR) was performed on a Rotor-Gene-2000 thermal cycler manufactured by Corbett Research using appropriate primers and 10 mcl of a PCR mixture (manufactured by “NPO Litech”) containing 2 mM MgCl2, Taq DNA polymerase and Cresol Red dye. The results were visualized by electrophoresis in 2% agarose gel with ethidium bromide at 150V and 290 mA. The size of the fragments was determined using the standard for the size of the lengths of DNA fragments from Life Technologies. To detect the studied polymorphisms, amplification of certain regions of the corresponding genes was performed. To determine the polymorphic alleles of cytokine genes, the method of restriction fragment length polymorphism (PDRF) was used.The polymorphism (C3435T) of the MDR1 gene was determined by PCR-PDRF. After the hot start and the first denaturation (94°C, 2 min), an amplification of 32 cycles was performed, each of which included: denaturation (94°C, 30 sec), annealing of primers (55°C, 1 min), elongation (74°C, 1 min). PCR was carried out in a mixture of 25 ml, which included 10 pmol of each primer, 200 microns of each dNTP, 1 ml (about 50 ng) of genomic DNA and a PCR mixture of the company "Interlabservice". 8.5 mcl of the PCR product with a length of 306 bp was incubated at 37°C with 0.5 mcl (3 units) of Ava-I restrictase and 1 mcl of a 10-fold restriction buffer. The restriction products were visualized by electrophoresis in a 2% agarose gel containing 1 mcg / ml of ethidium bromide. At the C3435T position of the MDR1 gene, the C-allele contains a restriction site and is detected by the presence of two fragments 190 and 116 bp, the T-allele does not contain a restriction site, the fragment is 306 bp. The polymorphism (C677T) of the MTHFR gene was determined by PCR-PDRF. After the hot start and the first denaturation (94°C, 10 min), an amplification of 35 cycles was performed, each of which included: denaturation (94°C, 10 sec), annealing of primers (55°C, 35 sec), elongation (72°C, 60 sec). PCR was performed in a mixture of 25 mcl, which included 8 pmol of each primer, 200 mcm of each dNTP, 1 mcl (about 50 ng) of genomic DNA and a PCR mixture of Interlabservice. 8.5 mcl of a PCR product 198 bp long was incubated at 37°C with 0.5 mcl (3 units) of Sali restrictase and 1 mcl of a 10-fold buffer for restriction. The restriction products were visualized by electrophoresis in a 2% agarose gel containing 1 mcg / ml of ethidium bromide. In the case of the presence of the allele-C, fragments of 140 and 58 bp were formed. In the presence of the allele-T, there is no restriction site, the fragment is 198 bp. Statistical processing of the results was carried out using the statistical software packages Arlequin 2006 (version 3.5.2.2), Excel 2003, SISA for calculating gene frequencies in the studied population.
Further purification of leukocyte mass lysates is based on the method of alcohol-salt treatment according to S. Miller et al. ((1988) in a modification proposed by the Stanford University Laboratory. To 400 ml of leukocyte mass lysate, 150 ml of 5M NaCl is added, mixed on a shaker and placed in ice for 10-20 minutes, then centrifuged at 1200 rpm for 15 minutes. The supernatant is taken into another Eppendorf-type test tube and 100% ice ethanol is added. With careful shaking, a quaternary chain of the DNA molecule appears in the mixture; the mixture is centrifuged at 1200 rpm for 15 minutes; the supernatant is removed, and the whitish spot remaining at the bottom of the test tube is washed again in 80% ethanol at 1200 rpm for 10 minutes. The supernatant is drained, the alcohol residues are carefully removed, and the test tube is left open until the alcohol completely evaporates (for 12 hours at room temperature or in a thermostat at a temperature of 40-45°C for 2 hours). After the alcohol has evaporated, a TE (Tris-EDTA) solution diluted with distilled water is added to the test tube with dried DNA in a ratio of 1:3 (TE: water) pH 8.0. The DNA was stored at – 20°C.Methods of identification of allelic variations of polymorphic loci of patients' genesPolymerase chain reaction (PCR) was performed on a Rotor-Gene-2000 thermal cycler manufactured by Corbett Research using appropriate primers and 10 mcl of a PCR mixture (manufactured by “NPO Litech”) containing 2 mM MgCl2, Taq DNA polymerase and Cresol Red dye. The results were visualized by electrophoresis in 2% agarose gel with ethidium bromide at 150V and 290 mA. The size of the fragments was determined using the standard for the size of the lengths of DNA fragments from Life Technologies. To detect the studied polymorphisms, amplification of certain regions of the corresponding genes was performed. To determine the polymorphic alleles of cytokine genes, the method of restriction fragment length polymorphism (PDRF) was used.The polymorphism (C3435T) of the MDR1 gene was determined by PCR-PDRF. After the hot start and the first denaturation (94°C, 2 min), an amplification of 32 cycles was performed, each of which included: denaturation (94°C, 30 sec), annealing of primers (55°C, 1 min), elongation (74°C, 1 min). PCR was carried out in a mixture of 25 ml, which included 10 pmol of each primer, 200 microns of each dNTP, 1 ml (about 50 ng) of genomic DNA and a PCR mixture of the company "Interlabservice". 8.5 mcl of the PCR product with a length of 306 bp was incubated at 37°C with 0.5 mcl (3 units) of Ava-I restrictase and 1 mcl of a 10-fold restriction buffer. The restriction products were visualized by electrophoresis in a 2% agarose gel containing 1 mcg / ml of ethidium bromide. At the C3435T position of the MDR1 gene, the C-allele contains a restriction site and is detected by the presence of two fragments 190 and 116 bp, the T-allele does not contain a restriction site, the fragment is 306 bp. The polymorphism (C677T) of the MTHFR gene was determined by PCR-PDRF. After the hot start and the first denaturation (94°C, 10 min), an amplification of 35 cycles was performed, each of which included: denaturation (94°C, 10 sec), annealing of primers (55°C, 35 sec), elongation (72°C, 60 sec). PCR was performed in a mixture of 25 mcl, which included 8 pmol of each primer, 200 mcm of each dNTP, 1 mcl (about 50 ng) of genomic DNA and a PCR mixture of Interlabservice. 8.5 mcl of a PCR product 198 bp long was incubated at 37°C with 0.5 mcl (3 units) of Sali restrictase and 1 mcl of a 10-fold buffer for restriction. The restriction products were visualized by electrophoresis in a 2% agarose gel containing 1 mcg / ml of ethidium bromide. In the case of the presence of the allele-C, fragments of 140 and 58 bp were formed. In the presence of the allele-T, there is no restriction site, the fragment is 198 bp. Statistical processing of the results was carried out using the statistical software packages Arlequin 2006 (version 3.5.2.2), Excel 2003, SISA for calculating gene frequencies in the studied population.3. Results
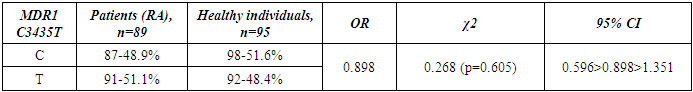
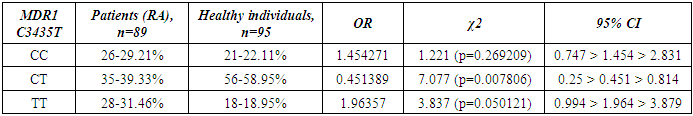
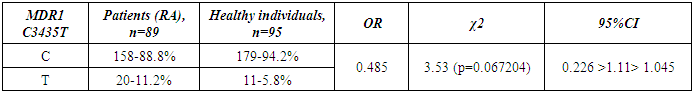
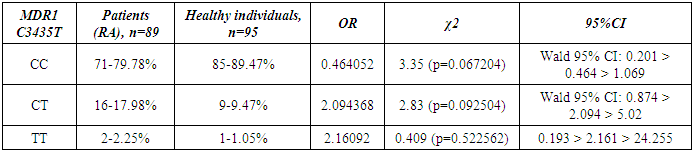
- Genotyping of the C3435T position of the polymorphic variant of the MDR1 gene was performed in a contingent of RA patients and healthy individuals of Uzbek ethnicity. The distribution of alleles of the polymorphic variant C3435T of the MDR1 gene in the group of RA patients and the control group included in the study is presented in Table 1. When studying the C3435T polymorphism of the MDR1 gene (multiple drug resistance gene) among healthy individuals, it was found that the frequency of the T-allele in the population of Uzbek people is 48.4%, the C-allele is 51.6%. When genotyping patients with RA, the T-allele was detected in 51.1% of cases, the C–allele in 48.9% of cases (χ2=0.268, p=0.268, OR=0.898). The analysis of the distribution of alleles in the group of RA patients compared with the control group of healthy individuals showed the absence of statistically significant differences in the polymorphism C3435T of the MDR1 gene – multiple drug resistance.
|
|
|
|
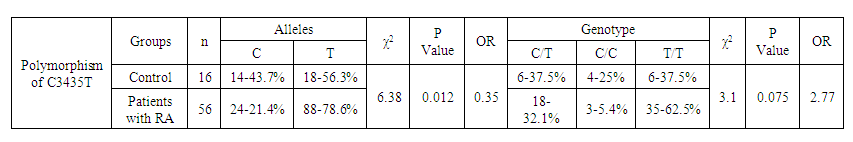 | Table 5. Distribution of frequencies of alleles and genotypes of the C3435T polymorphism of the MDR1 gene depending on the sex of RA patients and healthy individuals |
|
|
|
|
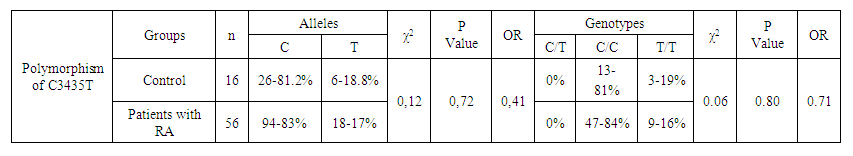 | Table 10. Distribution of frequencies of alleles and genotypes of C677T polymorphism of the MTHFR gene depending on the sex of patients with RA and healthy individuals |
4. Discussion
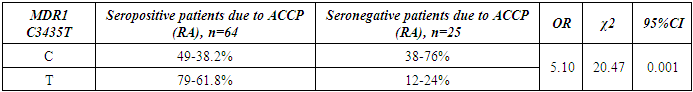
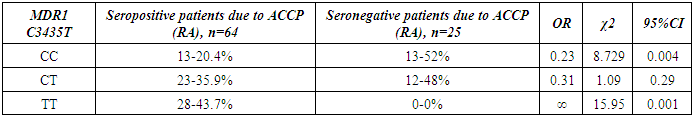
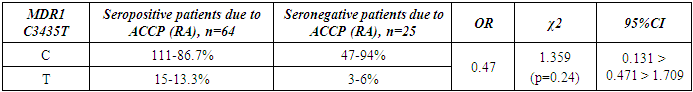
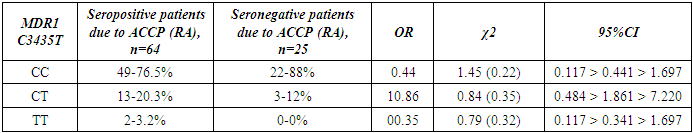
- The present study involved 117 patients with RA and 95 healthy people from the control group to assess the relationship between the 2SNP genes MDR1, MTHFR and susceptibility to RA. Our results showed that rs1045642 was significantly associated with RA, and rs1801133 polymorphism did not reveal significant associations with the RA.Pharmacogenetic studies show the relationship between genetic variability and drug response. The most important ones that can influence the therapeutic response to methotrexate are polymorphisms of the MDR1, MTHFR genes [15].The MDR1 gene belongs to the MDR family, localized on chromosome 7. The MDR family includes 2 human genes MDR 1 and MDR 2, but only MDR1 is related to multidrug resistance (MDR) [2]. The transcriptional activity of the gene increases in response to a variety of effects, including the effect of chemotherapeutic drugs. The association of some SNPs in the MDR1 gene with neurodegenerative diseases and diseases of the gastrointestinal tract was found [1,10]. Our studies confirmed the association of C3435T polymorphism of the MDR1 gene with RA, taking into account the gender and status of patients with ACСР.The MTHFR gene, located on the short arm of chromosome 1 (1p36.3), consists of 11 exons and 10 introns [12]. This gene encodes a 77 kDa protein [13]. MTHFR plays a role in the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate (5-methyl-THF). 5-methyl-THF acts as a methyl donor for remethylation of homocysteine to methionine. MTHFR polymorphisms play an important role in the response to treatment, as they affect the activity of enzymes and metabolism of methotrexate, among which are found: C677T (rs1801133) and A1298C (rs1801131) [14]. However, in our studies, no reliable association of polymorphism of the MTHFR gene (rs1801133) with rheumatoid arthritis was found. The current study had some limitations related to the sample size. Therefore, further studies are needed to confirm the association of the polymorphism of the MTHFR gene (rs1801133) with rheumatoid arthritis.
5. Conclusions
- Polymorphism C3435T (rs1045642) in the MDR1 gene is associated with RA. The allele T and the combination of genotypes C+T polymorphism MDR1 are markers of increased risk of RA. Polymorphism C677T in the MTHFR gene is not associated with RA in a sample of patients in the Uzbek population. An increase in the frequency of MDR1+3435TT and a decrease in the frequency of MDR1+3435CC genotypes were revealed in patients with seropositive form of RA compared with the seronegative subgroup. When comparing groups of patients based on the status of ACCP by polymorphic marker C677T in the MTHFR gene, no significant differences were revealed.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML