-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2020; 10(7): 445-447
doi:10.5923/j.ajmms.20201007.01

“Dry Drop” Method in the Practice of Genotyping and Determining the Presence of HIV-1 Drug Resistance Mutations in Uzbekistan
Nataliya G. Kan 1, Evgeniya Kazakova 2
1Reference Laboratory, Research Institute of Virology of MoH of Respublic of Uzbekistan, Tashkent, Uzbekistan
2Department of Molecular-Genetic Study, Research Institute of Virology of MoH of Respublic of Uzbekistan, Tashkent, Uzbekistan
Correspondence to: Evgeniya Kazakova , Department of Molecular-Genetic Study, Research Institute of Virology of MoH of Respublic of Uzbekistan, Tashkent, Uzbekistan.
| Email: |  |
Copyright © 2020 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Uzbekistan is a country with a large territory and a very hot climate, which creates additional difficulties in transporting native blood plasma from the regions to the laboratory conducting the research. In this regard, a component was included in the study on an alternative method of collecting biomaterial from patients and using the dry drop method for genotyping and determining the presence of mutations of HIV drug resistance. In parallel with the collection of native plasma in the three regions of the republic with the hottest climate, samples of the DPD and DBS were collected for HIV-1 drug resistance testing.
Keywords: HIV-1 drug resistance mutations, “Dry drop” method, Genotyping
Cite this paper: Nataliya G. Kan , Evgeniya Kazakova , “Dry Drop” Method in the Practice of Genotyping and Determining the Presence of HIV-1 Drug Resistance Mutations in Uzbekistan, American Journal of Medicine and Medical Sciences, Vol. 10 No. 7, 2020, pp. 445-447. doi: 10.5923/j.ajmms.20201007.01.
Article Outline
1. Introduction
- In 2014-15 on the territory of the Republic of Uzbekistan, the first national monitoring of HIV drug resistance was conducted according to the WHO recommendations [1,2]. The aim of the study was to determine the prevalence of HIV drug resistance among starting ART adult patients and among adult patients on ART. In 2005, the “dry drop” was first used in the Republic of Uzbekistan to determine HIV by ELISA [5]. Currently, in the republic every 2 years, sentinel surveillance of HIV infection is carried out using the “dry drop” method (PCR method is used to determine the proviral DNA). With the introduction into routine practice of new methods of laboratory diagnostics, it was decided to test the use of a “dry drop” for PCR and sequencing. Studies on PCR were carried out in 2013-14 [6].The dry drop method is attractive because of its availability. Sampling does not require clinical conditions (it is important when departing "on the field"); Does not require urgent additional processing; compact storage and less dangerous to handle; less chance of contamination; ease of obtaining, storage and transportation, samples of dry drops.
2. Overview
- Uzbekistan is a country with a large territory and a very hot dry climate, which creates additional difficulties in transporting native blood plasma from the regions to the laboratory conducting the research. In this regard, a component was included in the study on an alternative method of collecting biomaterial from patients and using the dry drop method for genotyping and determining the presence of mutations of HIV drug resistance. In parallel with the collection of native plasma in the three regions of the republic with the hottest climate, samples of the DPD and DBS were collected for testing.
2.1. Purpose of the Study
- The purpose of the study was to determine the prevalence of HIV drug resistance among starting ART adult patients and among adult patients on ART and compare methods for determining of HIV-1 drug resistance mutations.
2.2. Materials and Methods
- The study design was as follows: cross-sectional tracking with a duration of 7 months. The study involved all institutions providing ART in the Republic of Uzbekistan (15 centers to combat AIDS). The sample for each institution was proportional to the count of patients who started ART in 2014. The number of patients not previously ART expierence was calculated based on the number of patients registered in previous year and were randomly selected from them [3,4].During the period June-December 2015, 731 samples of native plasma were collected in the republic, of which ART-naive - 240 and 415 - were on ART with a recorded failure of treatment (HV ≥1000 copies). For many samples, the viral load were duplicated, since there was not always a fresh data of viral load over the past 6 months. In the three regions of the republic with the hottest climate, 43 samples of the DPD and DBS were collected for testing.The aim of the study was to adapt the method for both PCR and sequencing. Therefore, RNA isolation was carried out in three kits - Ribozol, RiboPrep and DNA-Sorb (all three sets of InterLabService production, Russian Federation).To obtain the eluate, lysing solutions of 3 above kits were used, as well as physical solution, and the incubation time was changed when the eluate was prepared.Extraction of RNA from the eluate was carried out according to the manufacturer's instructions for plasma. Further work on genotyping was carried out according to the instructions for the HIV AmpliSense HIV Genotyping Kit - AmpliSense HIV Genotype-Eph. At the same time, genotyping was performed using a standard procedure from blood plasma, as well as the determination of the HIV viral load.
3. Results and Discussion
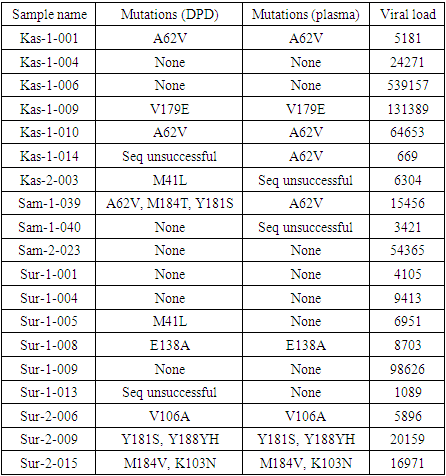
- 43 samples collected by the DPD and DBS methods, only 19 had a detectable viral load of HIV. Samples of DPD and DBS, as well as native plasma were obtained to genotyping to determine the presence of mutations of HIV drug resistance. The test results and detected drug resistance mutations are presented in the table. All paragraphs must be indented. All paragraphs must be justified alignment. With justified alignment, both sides of the paragraph are straight.
3.1. Results
|
3.2. Discussion
- When analyzing the results obtained, the following conclusions can be drawn. Of the analyzed sequences in 68.4% of cases, the comparison of the results obtained using plasma and the alternative method “dry drop” were identical. The use of the “dry drop” method with a viral load of at least 1000 copies of HIV RNA per ml is no less effective than the analysis of blood plasma.At last time there are publications that A62V is an additional mutation, which is often found in combination with mutations of resistance to NRTI K65R or Q151M [7]. A62V is widespread in the countries of the former Soviet Union, but otherwise A62 is non-polymorphic. The M184V / I mutation causes a high level of in vitro resistance to lamivudine and emtricitabine and a low level of didanosine and abacavir resistance [8,9,10]. However, M184V / I is not a contraindication to continue treatment with lamivudine or emtricitabine, since it increases susceptibility to zidovudine, tenofovir and stavudine and is associated with a clinically significant decrease in HIV-1 replication. The K103N mutation is a non-polymorphic mutation that causes a decrease in susceptibility to high-level nevirapine and efavirenz [11].Those, our study confirms the prevalence of this mutations, regardless of the chosen method of sample collection.
4. Conclusions
- When analyzing the results obtained, the following conclusions can be drawn. Of the analyzed sequences in 68.4% of cases, the comparison of the results obtained using plasma and the alternative method “dry drop” were identical. The use of the “dry drop” method with a viral load of at least 1000 copies of HIV RNA per ml is no less effective than the analysis of blood plasma.
ACKNOWLEDGEMENTS
- Many thanks to everyone who helped with the research, for the big support and trust to us.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML