-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2019; 9(3): 85-89
doi:10.5923/j.ajmms.20190903.04

Community-Acquired Carbapenemase-Producing Uropathogenic Enterobacteria Strains
Iskhakova H. I.1, Sapaeva F. R.1, А. Abdurakhimov A. A.2
1Tashkent Institute of Postgraduate Medical Education, Tashkent, Uzbekistan
2Institute of Biophysics and Biochemistry at the National University of Uzbekistan, Uzbekistan
Correspondence to: Sapaeva F. R., Tashkent Institute of Postgraduate Medical Education, Tashkent, Uzbekistan.
| Email: |  |
Copyright © 2019 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Microbiological analysis of urine was conducted at 426 patients of private diagnostic center «Vitras» diagnosed with acute uncomplicated urinary tract infection (UTI). Microbial growth in diagnostic titers (103 and above CFU/ml) was accounted for 38,7%, 147 (87,7%) of isolates related to enterobacteria, 112 (78.3%) from them related to E. coli. 15 strains with various phenotypes of resistance in DDT to the 3rd base carbapenemase (imipenem, meropenem, ertapenem) were investigated for carbapenemase-producing by using NordmannP acid-chemical method in modification of Pires J. et al. - CARBA/BLU method and in parallel by genic-molecular PCR method in «real time» mode. Identification of genes encoding of carbopenemase-producing was conducted by using 2 sets of «AmpliSens® MDRMBL-FL»: to identify the genes of acquired carbapenemase-producing by metal-β-lactamase (MBL) of the VIM, IMP, NDM groups and «AmpliSense® MDR» to identify KPC/OXA-48-FL produced by Federal Budget Institution of Science «Central Research Institute of Epidemiology» of The Federal Service on Customers' Rights Protection and Human Well-being Surveillance. At community-acquired E.coli uropathogenic strains all genes detected using by PCR (5 out of 15, 33,3%) related to metal-β-lactamase class. The most frequent NDM-1 gene was detected - at 3 - just NDM-1 emerged, at one strain together with VIM and one more strain of E.coli possessed only the VIM gene. All these strains with NDM-1 and VIM genes were positive in the phenotypic CARBA/BLU method. The IMP genes MBL class, КРС genes and OXA-48 genes were not found in the studied isolates. An accumulation of amplification products of the VIM gene and the OXA-48 gene at separate isolates of Escherichia with a positive CARBA/BLU reaction was observed later (after the 31st cycle).
Keywords: UPEC, MBL, NDM-1, OXA-48, VIM, IMP, CARBA/BLU, PCR
Cite this paper: Iskhakova H. I., Sapaeva F. R., А. Abdurakhimov A. A., Community-Acquired Carbapenemase-Producing Uropathogenic Enterobacteria Strains, American Journal of Medicine and Medical Sciences, Vol. 9 No. 3, 2019, pp. 85-89. doi: 10.5923/j.ajmms.20190903.04.
1. Introduction
- The infections of urogenital tract infections (UTI) occupy the first place among all urological diseases, they have been found in both outpatient and hospital practice, and Escherichia coli is currently considered the most significant uropathogenic bacteria [11 2, 14]. The resistance of Escherichia, Klebsiella, and other opportunistic pathogenic Gram-negative bacteria to antibiotics is one of urgent problems of public health all over the world [14, 4, 10]. A particularly important role is played by the resistance of these microorganisms to beta-lactam antibiotics; at the same time leading by mechanism of resistance for E.coli and other enterobacteria is ability to produce enzymes that inactivate beta-lactams [10, 6, 5, 9]. In addition to the extended-spectrum β-Lactamases (ESBLs) capable to hydrolysis of all cephalosporin’s, but not active on carbapenemase, all greater value is acquired carbapenemases active against all antibiotics of this class, excluding aztreonam [16, 10]. Carbapenems (imipenem, meropenem, ertapenem, doripenem) relate to reserve medicine; they have extensive coverage of action, low toxicity, good pharmacokinetic parameters, and the crucial - until recently a resistance to these medicine was rare enough [12, 13]. The occurrence and spread of carbapenemases-producing Enterobacteriaceae (CPE) and, in particular, Escherichia, is a significant negative factor for public health, therefore ensuring timely and reliable detection of CRE is the first crucial step in combating of this problem [4].The study goal was to study of community-acquired carbapenemase-producing uropathogenic Escherichia strains by phenotypic and genotypic methods.
2. Materials and Methods
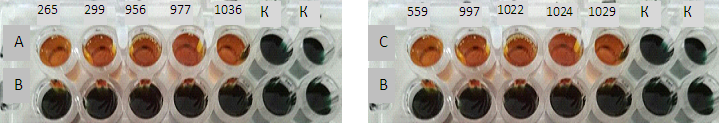
- The study was carried out at the Department of Microbiology of the Tashkent Institute of Postgraduate Medical Education; patients were admitted for examination by private diagnostic center «Vitras» LLC VITROSDIAGNOSTICS. 426 patients were diagnosed with a diagnosis of acute uncomplicated urinary tract infection (UTI) from October 2017 through March 2018. Cultivation of urine samples was carried out on Endo medium with selection of typical lacto-positive colonies. The study was carried out using by sectoral Gould method, microorganisms growing in urine dilution of 103 CFU/ml and above in accordance with International Standard [2]. Microbial growth in diagnostic titers was 38,7% (165 of 426), 147 (87,7%) of isolates related to enterobacteria, of which 112 (78.3%) related to E.coli. A sensitivity of the isolated Escherichia to various classes of antibiotics was determined by disk-diffusion test (DDT) according to recommendations of the EUCAST [15]. Internal quality control of determining the sensitivity to antibiotics was carried out with the verificatory Reference strain E.coli ATCC 25922. 15 strains of Escherichia with various resistance phenotypes in DDT to the 3rd basic carbapenems (imipenem, meropenem, ertapenem) were studied for carbapenemase-producing by NordmannP acid-chemical method in modification of Pires J. et al. by CARBA / BLU method and in parallel by genic-molecular PCR method. Genic-molecular testing was conducted in reference PCR laboratory of Diagnostics Research Institute of Virology of the Ministry of Health of Republic of Uzbekistan (Uzbekistan, Tashkent), in accordance with manufacturer's instructions of the set. Identification of genes encoding carbapenemase-producing was conducted using by 2 sets: «AmpliSens® MDRMBL-FL»: 1) to identify the genes of acquired carbapenemase-producing by metal-β-lactamase (MBL) of the VIM, IMP, NDM groups; 2) «AmpliSense® MDR» to identify KPC/OXA-48-FL produced by Federal Budget Institution of Science «Central Research Institute of Epidemiology» of The Federal Service on Customers' Rights Protection and Human Well-being Surveillance (Russia). Both sets are designed for PCR with hybridization-fluorescence detection of amplification products in "real time" mode. The PCR material was DNA samples obtained by extraction from samples of pure E.coli culture isolated from urine's patients with community-acquired UTI in titers of 103 / CFU / ml and above. Extraction of bacterial DNA was performed using by Ampli Prime DNA Sorb sets (Central Research Institute of Epidemiology, Russia) according to attached instructions. Amplification was carried out according to program in the tablet-type device.
|
3. Results and Discussion
- Considering the importance of antibiotic resistance problem, in order to determine the resistance of modern pathogens to antibiotics, great attention is paid to not only genic-molecular, but also to phenotypic methods that are available for implementation in routine practice. It is recommended (EUCAST) at the first stage to identify “suspicious” bacteria for the production of extended-spectrum β-Lactamases (ESBLs or carbapenemases) by ordinary DDT with 3 basic antibiotics of each group. When screening for carbapenemases, if at least one of the recommended carbapenems is stable (imipenem, meropenem and ertapenem), confirming the carbapenemase-producing can be validated by several phenotypic methods, including CARBA/BLU method. Earlier, we have found that local uropathogenic E.coli resistance to semisynthetic and inhibitor-protected penicillins and cephlosporins of the 2nd and 3rd generations was high, and majority (83,1%) of the studied cultures were classified as “suspicious” to ESBLs products, which was confirmed by disk combined method with clavulanic acid. It was also shown that resistance to carbapenems in screening with imipenem, meropenem and ertapenem in DDT in out-of-hospital E.coli averaged 59,1%, but phenotypes were different, the most frequent combination being sensitivity to imipenem, moderate resistance to meropenem and insensitivity to meropenem ertapenem. 15 strains were selected from total number of E. coli strains suspicious for carbapenemase-producing in DDT, and CARBA / BLU method was used as a confirmatory phenotypic method. These results presented in figure 1.
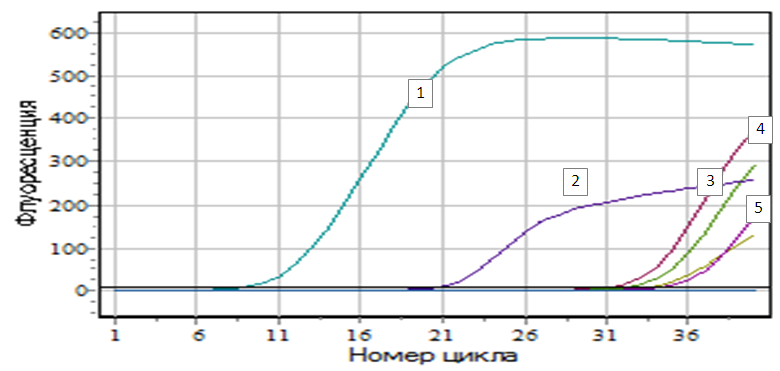 | Figure 2. Figure of accumulation of amplification products of NDM-1 genes: Legend: 1 - E.coli №956; 2 - K+; 3 - E.coli №1036; 4 - E.coli №977; 5- E.coli №265 |
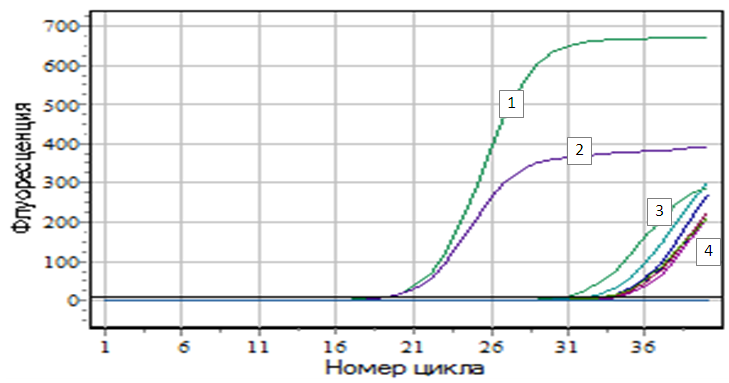 | Figure 3. Figure of accumulation of amplification products of VIM genes: Legend: 1 - K+; 2 - K+; 3 - E.coli №299; 4 - E.coli №956 |
4. Conclusions
- 1. In community-acquired uropathogenic E.coli strains, in 33,3% (5/15), metal-beta-lactamase genes were found. The most frequently detected NDM-1 gene - at 3 - only NDM-1; one strain in combination with VIM and one isolate - only VIM.2. All strains with the NDM-1 and VIM genes were positive in CARBA/BLU phenotypic method.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML