Jamoliddin Djololovich Khuzhakhmedov
Department of Molecular Medicine and Cell Technologies, Scientific Research Institute of Hematology and Blood Transfusion of the Ministry of Health of the Republic of Uzbekistan, Tashkent, Uzbekistan
Correspondence to: Jamoliddin Djololovich Khuzhakhmedov, Department of Molecular Medicine and Cell Technologies, Scientific Research Institute of Hematology and Blood Transfusion of the Ministry of Health of the Republic of Uzbekistan, Tashkent, Uzbekistan.
| Email: |  |
Copyright © 2018 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Abstract
The rates of himeric oncogene BCR/ABL р190, associated with translocation t (9; 22), were studied in patients with acute lymphoblastic leukemias (ALL) (34%). Hyperleukocytosis, thrombocytopenia, lymphadenopathy and hemorrhagic syndrome occurred in the onset of Ph - positive acute lymphoblastic leukemia.
Keywords:
Himeric oncogene BCR/ABL р190, Translocation, Acute lymphoblastic leukemia (ALL), Oncogene
Cite this paper: Jamoliddin Djololovich Khuzhakhmedov, Frequency of Acute Lymphoblastic Leukemia with Translocation t (9; 22) in Uzbekistan, American Journal of Medicine and Medical Sciences, Vol. 8 No. 1, 2018, pp. 329-332. doi: 10.5923/j.ajmms.20180811.06.
1. Introduction
It is known that the share of acute lymphoblastic leukemia (ALL) in adults is 20-25% [1, 5, 6], whereas in children this nosology occurs much more often – in 75-80% of cases of ALL [4]. Despite the success of modern polychemotherapy and the possibility of achieving complete remission in 80-85% of patients with ALL, long-term disease-free survival does not exceed 30-40% [9]. The most significant factors in the prognosis of leukemia are the presence of cytogenetic abnormalities. So to the most estimated prognostic factors of ALL are the translocations t (9; 22) and t (4; 11). The translocation t (9; 22) is defined in 5-10% of children and 25-30% of adult patients with ALL, and t (4; 11) is defined in children in 2%, in adults - in 5-6% of cases [3]. Identify the translocation of t (9; 22) by a standard cytogenetic method, the FISH method; as well as by polymerase chain reaction with reverse transcription (RT-PCR), which reveals the expression of the chimeric BCR/ABL gene [2, 8]. The detection of the chimeric BCR/ABL oncogene with ALL makes it possible to revise the therapeutic tactics, thereby increasing the duration of remission and the life expectancy of patients. However, in Uzbekistan to date, the frequency of occurrence of the chimeric BCR/ABL oncogene in ALL patients has not been studied, which was the reason for this study.
2. Main Body
2.1. Purpose of the Study
To study frequency of occurrence of the chimeric oncogene BCR/ABL in patients with ALL in Uzbekistan.
2.2. Material and Methods of Investigation
The study included 134 patients with a diagnosis of ALL who were on inpatient treatment at the Research Institute of Hematology and Blood Transfusion between January 2009 and December 2011. The age of the patients ranged from 15 to 65 years (median age 33.4 ± 3.1 years), of them men were 84 (63%), women – 50 (37%). The material of the study was blood / bone marrow, obtained from patients with ALL after admission to the hospital before the start of specific treatment.The fragments corresponding to the b3a2 type mRNA of BCR/ABL were 494 bp long, type b2a2 to 419 bp, type e1a2 to 502 bp. (Fig 1).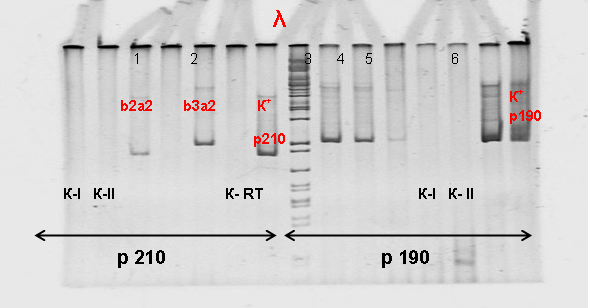 | Figure 1. Electrophoretic separation of the amplification products of the chimeric BCR/ABL gene in 6% PAAG. λ-marker of lengths; p210: b2a2 (419 bp), b3a2 (494 bp), p190: e1a2 (502 bp). K-RT-negative control of the RT reaction; K-I, K-II-negative control of PCR I and PCR II, respectively. № 1-6 samples of patients; K + p210 (b2a2), K + p190 (e1a2) |
The main criteria for the diagnosis of ALL were the general blood test, myelogram data and cytochemical study of blast cells. Patients were divided into 3 main groups. The first group included 102 patients with the first established ALL. The second group consisted of 16 patients with relapse of ALL. The third group consisted of 16 patients with primary-resistant course of ALL.In our study, RT-PCR was used to detect the chimeric BCR/ABL p190 and p210 transcript [7]. An important criterion for the diagnosis of Ph-positive ALL by the RT-PCR method was the double determination of a chimeric transcript, i.e. at its single revealing the result was considered negative.Statistical analysis of the results was carried out using a package of statistical programs "Microsoft Excel 2007". The following statistical parameters were calculated: the arithmetic mean, the arithmetic mean error, the value of the reliability criterion for differences (p) was checked using Student's t-test. The results of the statistical analysis were considered significant at p <0.05.
2.3. Results and Discussion
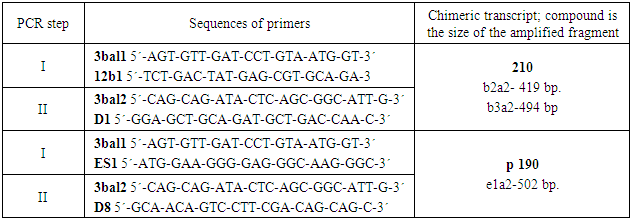
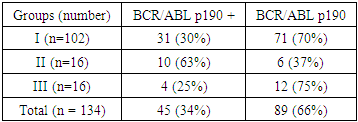
In the course of the study, it was found that expression of the BCR/ABL gene p190 was detected in 45 of 134 ALL patients, which was 34%. Table 1 shows the incidence of the oncogene BCR/ABL p190 in patients with 3 groups (Table 2).Table 1. Nucleotide sequences of primers used to detect chimeric BCR/ABL transcripts
 |
| |
|
Table 2. The occurrence of BCR/ABL p190 in patients with ALL
 |
| |
|
In Group I patients, the chimeric BCR/ABL p190 transcript was detected more than twice in 31 patients with ALL by RT-PCR, which accounted for 30% of all cases of ALL. In Group II patients, expression of the BCR/ABL p190 oncogene was detected in 10 patients, which was 63%. In the third group of patients, a chimeric transcript was detected in 4 patients, which was 25%.The control group in our study was 13 donors. Thirteen samples of peripheral blood were examined. The chimeric BCR/ABL p190 and p210 transcript was not detected by RT-PCR. In the course of the study, it was possible to establish some clinical and hematological patterns of the course of Ph-positive ALL. In the onset of the disease in patients with Ph-positive ALL, hyperleukocytosis was more common (the number of leukocytes was more than 50.0 × 109/l (Table 3). It was detected in 11 (35%) of 31 patients. In patients with Ph-negative ALL, hyperleukocytosis was registered in 21 (29%) of 71 cases. The aleukemic variant of the disease was more frequent in patients with Ph-negative ALL - in 19 (27%) cases out of 71. In the Ph-positive ALL, the aleukemic variant of leukemia was detected in 4 (13%) of 31 patients.Table 3 presents the main blood indices in Group I patients at the time of diagnosis of ALL.Table 3. Blood indicators of patients with Ph-positive ALL
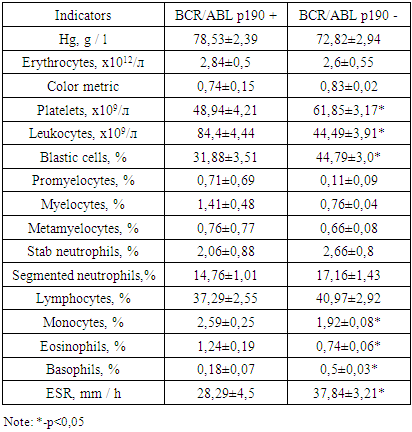 |
| |
|
As can be seen from Table 3, the amount of hemoglobin in the onset of the disease was not significantly different in ALL patients in the two subgroups and amounted to 78.53 ± 2.39 and 72.82 ± 2.94 g/l. The number of erythrocytes also did not differ significantly in both subgroups and amounted to 2.84 ± 0.5 and 2.6 ± 0.55 x 1012/l. The color index in patients with Ph-positive ALL was 0.74 ± 0.15, in patients with Ph-negative ALL – 0.83 ± 0.02, which did not differ significantly. The average number of leukocytes in patients with Ph-positive ALL in the onset of the disease was 84,4 ± 4,44х109/l, in patients without a pathological transcript - 44,49 ± 3,91х109/l. Thus, the number of leukocytes in the debut of the disease in patients with Ph-positive ALL was significantly higher by 1.9 times than in Ph-negative ALL (p<0.0001). At the time of diagnosis of Ph-positive ALL, the level of leukocytes varied widely: from 1.15x109/l to 800.0x109/l, with Ph-negative ALL, the spread of leukocytes was not so wide and amounted to 0.5-257.9x109/l. With respect to the average number of platelets, the opposite picture was observed. Thrombocytopenia was more pronounced in patients with Ph-positive ALL.The average number of platelets at the time of diagnosis was 48.94 ± 4.21x109/l, which is 1.26 times lower (p <0.05) relative to patients with Ph negative ALL, whose platelet count was 61.85 ± 3.17 x 109/l. The spread of platelets varied over a broader range in the ALL group without a pathological oncogene and amounted to 240 x 109/l, while in the subgroup with pathological oncogene it was 137 x 109/l. The average number of blast cells in the onset of the disease was lower in patients with Ph-positive ALL and was 31.88 ± 3.51%, while in patients with Ph-negative ALL - 44.79 ± 3.0% (p <0.05).The number of promyelocytes in the peripheral blood was statistically insignificant higher in the subgroup with Ph-positive ALL (0.71 ± 0.69%) relative to these patients with Ph-negative ALL (0.11 ± 0.09%) (p>0.05). The number of myelocytes in patients with pathological oncogene was is also statistically unreliable higher compared to patients without an oncogene and was 1.41 ± 0.48% compared to 0.76 ± 0.04% (p>0.05). The number of metamyelocytes in the two subgroups was approximately the same and amounted to 0.76 ± 0.77% and 0.66 ± 0.08%.Analysis of leucoformula showed the following changes. So the average number of neutrophils in the subgroups was different. The number of stab neutrophils in patients with Ph-positive ALL was 2.06 ± 0.88%, which was lower relative to patients with Ph-negative ALL, whose number was 2.66 ± 0.8%. The number of segmented neutrophils was statistically insignificantly higher (p> 0.05) than in patients without a pathological transcript (17.16 ± 1.43%) relative to patients with transcript expression (14.76 ± 1.01%). The average number of lymphocytes in the onset of the disease in patients with Ph-positive ALL was 37.29 ± 2.55%, in patients with Ph-negative ALL – 40.97 ± 2.92%, which does not differ significantly. The average number of monocytes in peripheral blood in patients with pathological oncogene was 1.3 times (p<0.02) higher than in patients without an oncogene and amounted to 2.59 ± 0.25% and 1.92 ± 0.08%, respectively. The number of eosinophils was also significantly higher 1.7 (p<0.02) times in patients with pathological transcript relative to patients without a pathological transcript, which amounted to 1.24 ± 0.19% compared to 0.74 ± 0.06% (p<0.001). The number of basophils, on the contrary, was 2.8 times higher in patients with Ph-negative ALL (0.18 ± 0.07%) relative to patients with Ph-positive ALL (0.5 ± 0.03%) (p<0,0001). The erythrocyte sedimentation rate (ESR) was statistically insignificant 1.3 times higher (p>0.05) in patients without oncogene expression and was 37.84 ± 3.21 mm/hr in patients with oncogene expression (28.29 ± 4.5 mm/h). Thus, in the onset of the disease in patients in two subgroups, a slight increase in ESR was recorded, which was due to an increase in the concentration of inflammatory protein markers (fibrinogen, C-reactive protein, etc.) in the plasma, as well as a decrease in the number of erythrocytes.Analysis of leucoformula showed the following changes. So the average number of neutrophils in the subgroups was different. The number of stab neutrophils in patients with Ph-positive ALL was 2.06 ± 0.88%, which was lower relative to patients with Ph-negative ALL, whose number was 2.66 ± 0.8%. The number of segmented neutrophils was 1.2 times higher (p<0.05) in patients without a pathological transcript (17.16 ± 1.43%) relative to patients with transcript expression (14.76 ± 1.01%). The average number of lymphocytes in the onset of the disease in patients with Ph-positive ALL was 37.29 ± 2.55%, in patients with Ph-negative ALL – 40.97 ± 2.92%, which does not differ significantly. The average number of monocytes in peripheral blood in patients with pathological oncogene was 1.3 times higher than in patients without an oncogene and amounted to 2.59 ± 0.25% and 1.92 ± 0.08%, respectively. The number of eosinophils was also significantly higher 1.7 times in patients with pathological transcript relative to patients without a pathological transcript, which amounted to 1.24 ± 0.19% compared to 0.74 ± 0.06% (p <0.001). The number of basophils, on the contrary, was 2.5 times higher in patients with Ph-negative ALL (0.18 ± 0.07%) relative to patients with Ph-positive ALL (0.5 ± 0.03%) (p <0.001). The erythrocyte sedimentation rate (ESR) was 1.4 times higher (p<0.01) in patients without oncogene expression and was 37.84 ± 3.21 mm/h in patients with oncogene expression (28.29 ± 4.5 mm/h). Thus, in the onset of the disease in patients in two subgroups, an increase in ESR was recorded, which was due to an increase in the concentration of inflammatory protein markers (fibrinogen, C-reactive protein, etc.) in the plasma, as well as a decrease in the number of erythrocytes.With careful collection of anamnesis and objective examination of patients with Ph - positive ALL, 59% of cases were detected, 47% of cases of intoxication syndrome, 35% of hemorrhagic syndrome, 29% of splenomegaly, 24% of immunodeficiency syndrome, 18% of anemic syndrome as shown in Figure 2.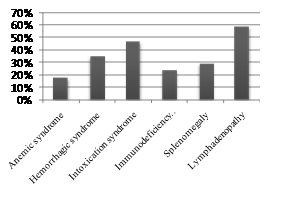 | Figure 2. Frequency of occurrence of syndromes in primary patients with Ph-positive ALL |
As can be seen from Figure 1, most often Ph-positive ALL debuts with an increase in peripheral lymph nodes, intoxication syndrome and hemorrhagic syndrome. Lymphadenopathy was characterized by an increase in tonsils and lymph nodes both peripheral and internal (intrathoracic and intra-abdominal) to various sizes, painless for palpation. Hemorrhagic syndrome manifested itself from small-scale petechiae and ecchymosis to external bleeding (most often nasal and female uterus). The development of hemorrhagic syndrome was due to two main factors - a critical decrease in platelets and a violation of the vascular wall nutrition due to platelet dysfunction.
3. Conclusions
The frequency of occurrence of the chimeric oncogene BCR/ABL p190 in patients with ALL in Uzbekistan was 34%.Ph - positive ALL often debuts with hyperleukocytosis, expressed thrombocytopenia, left leukocyte shift, lymphadenopathy and hemorrhagic syndrome.The chimeric transcript of BCR / ABL p190 and p210 was not detected by RT-PCR among donors in the control group, and BCR / ABL p210 was not detected among patients with ALL.
ACKNOWLEDGEMENTS
The author thanks the scientific adviser and the head of the Department of Molecular Medicine and Cell Technologies of, Scientific Research Institute of Hematology and Blood Transfusion of the Ministry of Health of the Republic of Uzbekistan Hamid Yakubovich Karimov and the head of the laboratory of medical genetics Kodirzhon Tukhtabaevich Boboev for assistance in the management and organization of the conducted studies.
References
| [1] | Vorobiev A. I. Manual on hematology. In 3 volumes - Moscow: Nyudiamed, 2007. T.1. 1275 p. |
| [2] | Domracheva E. V., and Aseeva E. A. Possibilities and prospects of hematological cytogenetics // Honey. Genetics. - 2004. - №4. - pp. 166-179. |
| [3] | Parovichnikova E. N., Mavrina E. S., Surin V. L. with et al. Methods of monitoring the minimal residual disease in patients with acute lymphoblastic leukemia // Hematol. and transfuziol. - 2013. - №3. - P. 45-48. |
| [4] | Rukavitsyn O.A. Hematology: national leadership. - Moscow.: GEOTAR-Media, 2015. - 776p. |
| [5] | Savchenko V. G., Galtseva I. V., Parovnikikova E. N.. Program treatment of leukemia. - Moscow: Foliant, 2008. – 484 p. |
| [6] | Turkina AG, Zarytsky A.Yu., Shuvaev V.A. with et al. Clinical recommendations for the diagnosis and treatment of chronic myelogenous leukemia // Clinical oncohematology - 2017. - No. 3. - P. 294-316. |
| [7] | Ottmann O.G., Pfeifer H. First-line treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia in adults // Cur. Opin. Oncol. – 2009. – v. 21. - Р. S43-S46. |
| [8] | Kurzrock R., Kantarjian H. M., Druker B. J. et al. Philadelphia chromosome-positive leukemias: from basic mechanisms to molecular therapeutics // Ann. Intern. Med. – 2003. – v. 138. – Р. 819-830. |
| [9] | Thomas X. Philadelphia chromosome-positive acute lymphoblastic leukemia stem cells in acute lymphoblastic leukemia and tyrosine-kinase inhibitor therapy // World J. Stemm. Cells. - 2012. – v. 26. – P. 44-52. |



 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML


