-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2015; 5(5): 231-234
doi:10.5923/j.ajmms.20150505.08
Endothelial Nitric Oxide Synthase Gene Polymorphism (T-786 C) in Sudanese Patients with Sickle Cell Anaemia
Nada H. Eltayeb1, 2, Mohamed A. M. Salih3, Abdel Rahim M. Muddathir4, 5
1Department of Haematology, Faculty of Medical Laboratory Sciences, AlNeelain University, Khartoum, Sudan
2Alneelain Medical research centre, AlNeelain University, Khartoum, Sudan
3Central laboratory, Ministry of Science and Technology, Khartoum, Sudan
4Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, Taibah University, Almadina Almunawarah, Kingdom of Saudi Arabia
5Department of Haematology and Blood Transfusion, Faculty of Medical Laboratory Sciences, Alzaiem Alazhari University, Khartoum North, Sudan
Correspondence to: Abdel Rahim M. Muddathir, Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, Taibah University, Almadina Almunawarah, Kingdom of Saudi Arabia.
| Email: |  |
Copyright © 2015 Scientific & Academic Publishing. All Rights Reserved.
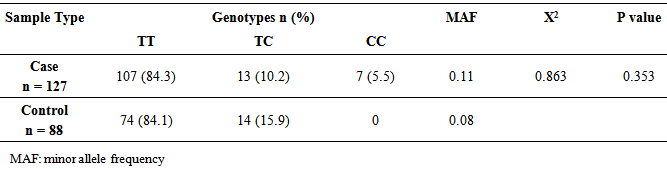
Background and Objective: Endothelial nitric oxide synthase gene polymorphisms have been associated with the sickle cell anemia. The aim of conducting this study was to explore the association between the single nucleotide polymorphism (T-786C) of endothelial nitric oxide synthase gene and Sickle cell anaemia among Sudanese patients. Method: This was a case control study in which a total of 127 patients with sickle cell disease and 88 Healthy controls were involved. The single nucleotide polymorphism (T-786C) was genotyped by Polymerase Chain Reaction – Restriction Fragment Length Polymorphism and the association was tested by Haploview software. Result: The distribution of mutant allele -786C was present in 5.5% of patients and completely absent in control subjects, but there is no significant association between single nucleotide polymorphism (T-786C) and Sickle cell Disease in Sudan (p>0.05). Conclusions: This study indicated lack of association between single nucleotide polymorphism (T-786C) and Sickle cell Disease among Sudanese patients.
Keywords: Endothelial Nitric Oxide Synthase, T-786C, Polymorphism, Sickle Cell Anaemia
Cite this paper: Nada H. Eltayeb, Mohamed A. M. Salih, Abdel Rahim M. Muddathir, Endothelial Nitric Oxide Synthase Gene Polymorphism (T-786 C) in Sudanese Patients with Sickle Cell Anaemia, American Journal of Medicine and Medical Sciences, Vol. 5 No. 5, 2015, pp. 231-234. doi: 10.5923/j.ajmms.20150505.08.
Article Outline
1. Introduction
- Sickle cell anaemia is characterized by a high variable clinical course [1]. The presence of HBB (Human beta-globin gene) gene product, sickle hemoglobin (HbS:α2 βS2), alone is not enough to cause the variability in clinical course of the Sickle Cell Disease [2]. Phenotypic heterogeneity of the Sickle Cell Disease (SCD) with different clinical outcomes seem to be modulated by polymorphisms in genes that are involved in inflammation, cell–cell interaction and modulators of oxidant injury and nitric oxide (NO) biology [1]. NO has properties that can impact every aspect of SCD, from decreasing platelet activation and adhesion receptor expression on the vascular endothelium, to decreasing vascular smooth muscle proliferation, limiting ischemia-reperfusion injury, modulating endothelial proliferation, and regulating inflammation [3]. In vivo NO is synthesized during the enzymatic conversion of L-arginine to L-citrulline by three isoforms of nitric oxide synthase (NOS) enzyme, namely, neuronal NOS (nNOS or NOSI), inducible NOS (iNOSor NOSII), and endothelial NOS (eNOS or NOSIII). [4] Endothelial (e) NOS, derived from vascular endothelium, is the most dominant form of these isoforms [5].The eNOS is encoded by a gene located on chromosome 7q35-q36, which is 21kb in size and consists of 26 exons [6, 7]. Additionally, promoter region of the eNOS gene harbors several transcription factor binding sites, regulating gene expression [8]. The level of NO in the body is linked to expression of eNOS gene [9]. The Single Nucleotide Polymorphism (SNP) (T-786C) (rs2070744) in the 5. promoter region affects the expression of eNOS gene. The -786C allele binds the inhibitory transcription factor protein A1 resulting in a low mRNA level of eNOS and this reduces NO production and endothelial function [10].Reduced endothelial NO bioavailability in SCD leads to activation of endothelial cell adhesion molecules besides platelet activation, which resulting into vascular occlusion and vaso constriction. [11]Many published reports have suggested the involvement of eNOS polymorphisms in the pathogenesis of sickle cell complications such as acute chest syndrome and painful vasoocclusive crises [12 – 15]. There is a substantial interethnic diversity in the distribution of eNOS variants [16-20], and this difference could potentially clarify interethnic differences in nitric oxide bioavailability and potentially sickle cell pathophysiology [21]. In Sudan, Sickle cell anaemia presentation was severe and it was frequently fatal in early childhood and was accompanied with major complications [22-25]. The aim of conducting this study was to investigate the association of the eNOS SNP (T-786C) with SCD among Sudanese patients.
2. Materials and Methods
- This prospective case control study in which a total of 127 confirmed Sicklerpatients, their age ranged 6 months to 42 years old mean (10.8±7.1, sd) attending Jaafar Ibn Oaf Hospital and Military Hospital for regular checkup, in addition to 88 Healthy controls were involved. Informed consent obtained from all participants and the study was approved by Ministry Health of Khartoum. .Clinical and demographic data were collected in a predesigned questionnaire.DNA Extraction Human genomic DNA was extracted from blood samples using a simple salting-out method [26].Genotyping of T-786C polymorphismThe SNP (T-786C) in the 5’ promoter region of the eNOS gene has been determined byPolymerase Chain Reaction – Restriction Fragment Length Polymorphism (PCR-RFLP) analysis using forward primer5’GAGTCTGGCCAACACAAATCC3’ and reverse primer 5’ACCTCTAGGGTCATGCAGGT3’. PCR reaction mixture was prepared by adding 5 μlgenomic DNA to a total volume of 20 μlready master mix (Maxime PCR PreMix Kit (i-Taq), iNtRON biotechnology). Initially DNA was denatured at 94°C for 2minutes. PCR amplification protocol was: Thirty cycles, consisting of 20seconds denaturation at 94°C, 10 seconds annealing at 58.5°C, and 40 seconds extension at 72°C and the final extension included a 3.5minutes extension at 72°C. The PCR fragment (658 bp) was digested with 1 unit of Hpa II restriction enzyme at 37°C. The digested and undigested PCR products were estimated with 5 μL 100 bp DNA ladder in 1.5% agarose gels. Gel image was captured by Gel Documentation System (SYNGENE, JAPAN) and analyzed using the Image Analysis Software (GeneSnap 7). According to Webcutter 2.0 program(rna.lundberg.gu.se/cutter2) homozygous wildallele (TT) produced 2 fragments 374 bp and 284 bp; heterozygous mutant allele (TC) produced 3 fragments 374 bp, 328 bp, and 284 bp; whereas homozygous mutants allele (CC) produced 2 PCR fragments 328 bp and 284 bp (figure 1). Ten percentof the samples subjected to repeat PCR and genotyping for quality control purposes and the results were found to be 100% concordant.
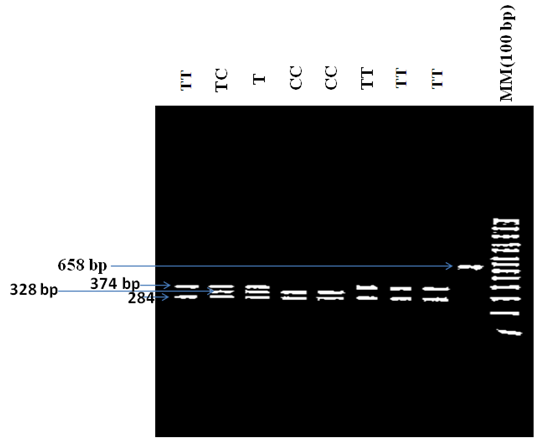 | Figure 1. Agarose gel electrophoresis Showering variants of the T-786C (rs 2070744) polymorphism of the endothelial nitric oxide synthasegene. bp: base pair |
3. Result
- The study results showed that, there was higher frequency of wild type (TT) followed by heterozygous allele (TC) among both patients and controls; whereas mutant allele (CC) present in patients and completely absent among control subjects (table 1). Genotype and allele frequencies of eNOS SNP (T-786C) showed no statistical significance of differences between the sickle cell disease and controls (χ2 = 0.863, P=0.353). The associations between eNOS SNP (T-786C) and SCD tested by Haploview software showed no associations observed for eNOS SNP (T-786C) with SCD in Sudanese patients. The genotype distribution for the eNOS SNP (T-786C) in Sudanese SCD population was in line with Hardy–Weinberg equilibrium (P=1.0).
|
4. Discussion
- In order to verify the role of the eNOS gene polymorphisms in sickle cell anaemia and to confirm the effect of interethnic diversity in the distribution of eNOS variants, we performed a case-control study and genotyped eNOS SNP (T-786C) in 127 cases of SCD and 88 controls. To our knowledge this is the first study to examine the role of the eNOSSNP (T-786C) in the pathogenicity of sickle cell anaemia in Sudan. Many published reports have suggested the involvement of eNOS polymorphisms in the pathogenesis of sickle cell complication [12-15]. Tanus-Santos et al found marked interethnic differences in the distribution of eNOS variants: (-786C) allele was more common in Caucasians than in Asians or African-Americans [16]. Few published reports worldwide are available concerning the association of eNOS SNP (T-786C) with sickle cell disease. A study from Mali reported no significant difference in the frequency of mutant allele (-786C) of eNOS gene between patients and controls (2.3% and 2.2% respectively) [21]. Also another study from Brazil showed no significant difference in the frequency of mutant allele (-786C) of eNOS gene between patients and controls (0.05% and 0.06% respectively) [28]. While a study from India reported significant difference in the frequency of mutant allele (-786C) between SCD and control groups (16.6% and 1.33% respectively) [29]. The present study showed no associations between the eNOS SNP (T-786C) and sickle cell anaemia in Sudan, so our study was in agreement with a study done by Thakur et al from Mali, which reported no significant difference in the genotypic frequencies of eNOS (T-786C) polymorphism between sickle cell disease and controls [21]. Also our finding in agreement with another Study from Brazil which indicated lack of association of eNOS SNP (T-786C) with sickle cell disease [28]. In contrastour finding in disagreement from study done in India that showing a significant association of eNOS SNP (T-786C) with SCD [29]. Factors such as sample sizes, ethnic heterogeneity and environmental factors contributed to this contrast in results.
5. Conclusions
- The present study showed no associations between the eNOS SNP (T-786C) and sickle cell anaemia in Sudan. Additional functional studies on eNOS gene polymorphisms might help understanding of the physiopathology of the disease and permitprognostic and therapeutic advances in the area of sickle cell disease.
ACKNOWLEDGMENTS
- We thank Miss Awatif Salahelden Suliman for her technical support.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML