-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Medicine and Medical Sciences
p-ISSN: 2165-901X e-ISSN: 2165-9036
2012; 2(1): 20-24
doi: 10.5923/j.ajmms.20120201.05
Antibiotic Resistance Profile of Gram Negative Bacteria Isolated from Surgical Wounds in Minna, Bida, Kontagora and Suleja Areas of Niger State
Sani R. A. 1, Garba S. A. 2, Oyewole O. A. 2
1Hospital Management Board, Minna, Niger State, Nigeria
2Department of Microbiology, Federal University of Technology Minna, Niger State, Nigeria
Correspondence to: Oyewole O. A. , Department of Microbiology, Federal University of Technology Minna, Niger State, Nigeria.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Antibiotics resistance profiles of bacteria from surgical wounds were investigated in four (4) General Hospitals (Bida, Kontagora, Minna and Suleja) in Niger State. Five hundred (500) samples (i.e. Two hundred (200) samples in Minna, One hundred (100) samples each Suleja, Kontagora and Bida) of wound exudates from these general hospitals, were analysed. The results showed the presence of Klebsiella Pneumoniae, Escherichia coli, Pseudomonas aeruginosa, and Proteus vulgaris in the samples from the wounds. From the five hundred (500) samples collected from all locations, forty two (42) samples had Kl. pneumoniae, sixty four (64) samples had Ps. aeruginosa, fifty two (52) samples had P. vulgaris and one hundred and nine (109) samples had E. coli. E. coli was the most frequently isolated bacteria from wounds in all the locations, while Kl. pneumoniae was the least isolated from wounds in all the locations. All the bacteria were tested for sensitivity against tarivid, pefloxacin, ciprofloxacin, augmentin, gentamycin, streptomycin, ceporex, nalidixic acid, septrin, ampicillin, ampiclox, zinacef, amoxacillin, rocephin, erythromycin. Most of all the isolates were sensitive to ciprofloxacin, pefloxacin and Tarivid while others were resistant to remaining antibiotics. E. coli, Strept. pyogenes and S. aureus showed highest resistance profile and P. vulgaris, Kl. pneumoniae and Ps. aeruginosa showed least resistance profile to most antibiotics used.
Keywords: Antibiotic, Resistance, Sensitive, Surgical Wound
Cite this paper: Sani R. A. , Garba S. A. , Oyewole O. A. , "Antibiotic Resistance Profile of Gram Negative Bacteria Isolated from Surgical Wounds in Minna, Bida, Kontagora and Suleja Areas of Niger State", American Journal of Medicine and Medical Sciences, Vol. 2 No. 1, 2012, pp. 20-24. doi: 10.5923/j.ajmms.20120201.05.
Article Outline
1. Introduction
- Wound is defined as an injury to any of the tissues of the body, especially the one that is caused by physical means and with interruption of continuity (Giacometti et al., 2000). The most common underlying event for all wounds is trauma. Trauma may be accidental or intentionally induced. The intentionally induced trauma category includes hospital – acquired wounds, which can be grouped according to how they are acquired, such as surgically and by use of intravenous medical devices. Although, none intentionally induced, hospital-acquired wounds can be the pressure sores caused by ischemia. They are also referred to as decubitus ulcers (bedsore), and when such wounds become infected, they are often colonizing bacterial species (Giacometti et al., 2000).Wound can be infected by a variety of microorganisms ranging from bacteria to fungi and parasites (Bowler et al., 2001). The common gram positive organisms are the β –hemolytic Streptococcus – Strept. pyogenes and S. aureus.The gram negative aerobic rods are Ps. aeruginosa. The facultative anaerobes include Enterobacter species, E. coli, Klebsiella species and Proteus species. The fungi are Candida species and Aspergillus species (Gus Gunzalez et al.,2006; Mordi and Momoh, 2009). The control of wound infections has become more challenging due to widespread bacterial resistance to antibiotics and to a greater incidence of infections caused by Methicillin – resistant S. aureus (MRSA) and Vancomycin resistant Enterobacter (VRE), polymicrobic flora and by fungi.The knowledge of the causative agents of wound infection has proved to be helpful in the selection of empiric antimicrobial therapy and on infection control measures in hospitals (Shittu et al., 2004) are also useful in formulating rational antibiotic policy. The aim of this research therefore is to determine the antibiotic resistance profile of gram negative bacteria isolated from surgical wounds in Minna, Bida, Kontagora and Suleja areas of Niger State, Nigeria.
2. Materials and Methods
2.1. Collection of Samples
- Wounds samples were collected from five hundred (500) patients that undergo surgical operation in four (4) general hospitals in Minna, Bida, Kontagora and Suleja areas of Niger State. 200 samples were collected from general hospital in Minna while 100 samples were collected each from Bida, Kontagora and Suleja general hospitals. The wound types included boils, whitlow, abscesses, cervicitis, trauma wounds, burns, systemic ulcers, insect bites and swelling of no specific etiology. These samples were transferred to the Microbiology laboratory of Federal University of Technology, Minna, Niger State, Nigeria for further analysis.
2.2. Characterization and Identification of the Isolates
- The collected samples were streaked on freshly prepared nutrient agar plates and incubated aerobically and anaerobically at 37oC for 24 hours. Bacterial colonies differing in size, shape and colour were selected from the different plates and further subcultured on nutrient agar by the streak plate technique and incubated at 37oC for 24 hours after which, were maintained in agar slants for further characterization and identification. The bacterial isolates were characterized based on colonial and cell morphology, growth on differential/selective media and biochemical tests which include Gram’s reaction, indole tests, methyl red, voges-proskauer, citrate utilization, motility, endospore, utilization of carbohydrates such as glucose, sucrose, mannitol, lactose and fructose, oxidase, catalase, coagulase and starch hydrolysis test (Oyeleke and Manga, 2008). The bacterial isolates were identified by comparing their characteristics with those of known taxonomy using the schemes of Cowan and Steel (1993).
2.3. Susceptibility of Isolates to Various Antibiotics
- Antibiotic sensitivity tests were carried out on all isolates using paper (New Man England) disc diffusion technique. A total of 10 antibiotics were tested and 0.2ml of 12h peptone water culture of test organism was used to inoculate each organism on a dry sterile nutrient agar plate. The resistant profiles of bacteria isolated from surgical wounds were determined by standard methods.The antibiotic discs used were gram negative sensitive as follows: tarivid, pefloxacin, ciprofloxacin, augmentin, gentamycin, streptomycin, ceporex, septrin, ampicillin. Nutrient agar was the media used. Each of the isolates was spread over the entire surface of the nutrient agar using a sterile glass spreader and allowed to dry for about 15 to 30min. The antibiotic discs were placed on agar using sterile forceps. The plates with the antibiotic discs were then incubated at 37oC for 24 hours to observe the zones of growth inhibition produced by the antibiotics and recorded immediately.
3. Result
3.1. Microorganisms Isolated from Samples at Each Location
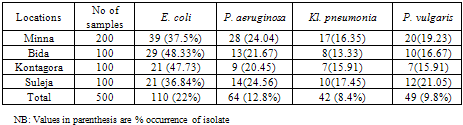
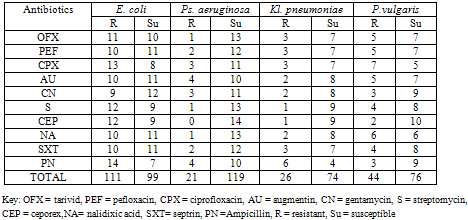
- Table 1 shows the gram negative bacteria isolated from wound samples in various general hospitals examined. E. coli had the highest occurrence in all four locations (22%) followed by P. aeruginosa (12.8%) while Kl. pneumonia had the least occurrence (8.4%).
3.2. Antibiotic Resistance of Gram Negative Bacteria in Minna
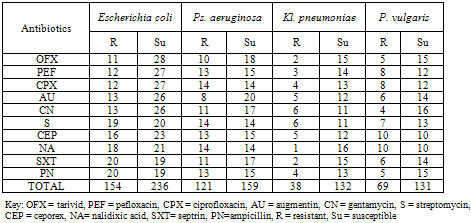
- Table 2 shows the antibiotics resistance of gram negative bacteria isolated from General hospital Minna. E. coli was most resistant while Kl. pneuomoniae was least resistant to the antibiotics examined while E. coli was most susceptible to all the antibiotics and P. vulgaris was most susceptible to all the antibiotics.
3.3. Antibiotic Resistance of Gram Negative Bacteria in Bida
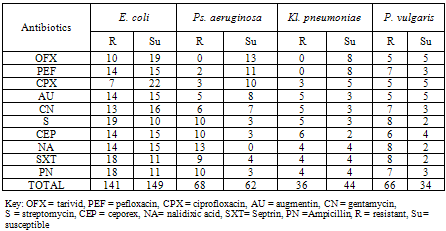
- Table 3 shows the antibiotics resistance of gram negative bacteria isolated from General hospital Bida. E. coli was most resistant while Kl. pneumoniae was least resistant to the antibiotics examined while E. coli was most susceptible to all the antibiotics and P. vulgaris was most susceptible to all the antibiotics.
3.4. Antibiotic Resistance of Gram Negative Bacteria in Kontangora
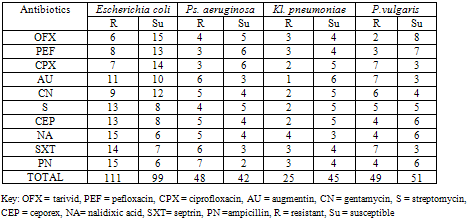
- Table 4 shows the antibiotics resistance of gram negative bacteria isolated from General hospital Kontagora. E. coli was most resistant while Kl. pneuomoniae was least resistant to the antibiotics examined while E. coli was most susceptible to all the antibiotics and Ps. aeruginosa was most susceptible to all the antibiotics.
3.5. Antibiotic Resistance of Gram Negative Bacteria in Suleja
- Table 5 shows the antibiotics resistance of gram negative bacteria isolated from General hospital Kontagora. E. coli was most resistant while Ps. aeruginosa was least resistant to the antibiotics examined while Ps. aeruginosa was most susceptible to all the antibiotics and Kl. pneumoniae was most susceptible to all the antibiotics.
4. Discussion
- The gram negative organisms isolated in wound infection were E. coli, P. vulgaris, Ps. aeruginosa and Kl. pneumoniae. Of all the organisms isolated, E. coli had the highest frequency of occurrence (22%) followed by Ps. aeruginosa (12.8%). This finding was in agreement with Siguan et al. (1987) and Olayinka et al. (2004) which stated that the common organisms isolated from surgical wound were gram-negative microbes comprising P. aeruginosa, E. coli and Enterobacter spp.E. coli showed the highest resistance to the antibiotics considered in all the locations. This was in agreement with the reports of Olayinka et al. (2004) which stated that E. coli was the commonest agent of bacteraemia and the one showing the most striking changes in resistance, especially to septrin and ampicillin. High incidence of E. coli in wound infection was reported by Wazait et al. (2003). The alarming trend is the speed with which aminoglycoside and penicillin resistance is accumulating in E. coli all over Africa especially in Nigeria (Hefferman and Woodhouse, 2007). Besides, E. coli easily acquire resistance factor from environment and they are easily resistant to penicillin derivatives drug like ampicillin and oxacillin (Wazait et al., 2003).Kl. pneumoniae showed resistance to all the antibiotics except tarivid and pefloxacin, in Bida. Ps. aeruginosa was resistant to ceporex in all locations except in Suleja. P. aeruginosa also showed no resistant to tarivid in Bida. E. coli, Ps. aeruginosa and Kl. pneumoniae can acquire resistance plasmid in a mixed culture. This explains further why most of these organisms are resistant to antibiotics (Nester et al., 2004).Proteus vulgaris is most frequently recovered from immunocompromised patients or those on long-term antibiotic regimen. This organism was resistant to all the antibiotics in all the locations. This finding was contrary to the reports of Mordi and Momoh (2009) which stated that P. vulgaris was sensitive to all antibiotics except chloramphenicol.Klebsiella pneumoniae was the least commonly isolated organisms amongst the Gram negative facultative anaerobic bacilli. This is however contrary to the observations of Kehinde et al. (2004) who claimed that Kl. pneumoniae was most predominant in surgical wounds. But this finding was in line with the reports of Stock and Wiedemann (2001) which stated that Kl. pneumoniae was the least isolated in surgical wounds.
|
|
|
|
|
5. Conclusions
- The findings of this study suggest that bacterial resistance in surgical wound infections is becoming serious menace in all the locations/study area.E. coli is still the most frequently involved pathogen, showing high resistance rates of bacteria isolated from surgical wounds.Tarivid, ciprofloxacin and Pefloxacin are the best therapeutic options to treat these infections because of the resistant caused by these organisms.Infections of the surgical wound by these bacteria are one of the most common and important cause of morbidity and mortality. The delay in recovery and subsequent increased length of hospital stay also has economic consequences. It has been estimated that each patient with a surgical site infection will require an additional six to seven (6-7) days in the hospital, which results in the doubling of hospital costs. Early treatment: when antibacterial therapy is indeed necessary, it should be promptly initiated; inadequate use of antibacterial (e.g., doses that are too low, therapy ended prematurely) is a major factor for the selection of resistant strains
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML