-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Intelligent Systems
p-ISSN: 2165-8978 e-ISSN: 2165-8994
2012; 2(4): 66-75
doi: 10.5923/j.ajis.20120204.06
Genotype Division for Shared Memory Parallel Genetic Algorithm Across Platforms and Systems
Dana Vrajitoru
Computer and Information Sciences Department, Indiana University South Bend, South Bend, IN 46545, USA
Correspondence to: Dana Vrajitoru , Computer and Information Sciences Department, Indiana University South Bend, South Bend, IN 46545, USA.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
In this paper we present a concurrent implementation of coevolutionary genetic algorithm (GA) designed for shared memory architectures such as multi-core processor platforms. Our algorithm divides the chromosome among the processes, and not the population as it is the case for most parallel implementations of the GA. This approach results in a division of the problem to be solved by the GA into sub-problems. We analyze the influence on performance and speedup of several parameters defining the algorithm, such as: a synchronous or asynchronous information exchange between processes and the frequency of communication between processes. We also examine how the problem separability influences the general algorithm performance. Finally, we compare different operating systems and platforms in the evaluation process. Our paper shows that this approach is a good way to take advantage of multi-core processers and improve not only the execution time, but also the fitness in many cases.
Keywords: Genetic Algorithms, Shared Memory Models, Genotype Division
Article Outline
1. Introduction
- The hardware developments from recent years have made multi-core architectures a common place in the industry. The issue we are now facing is taking advantage of such platforms.Parallel and distributed versions of the genetic algorithms (GAs) are popular and diverse. The simplest parallel models are function-based where the evaluation of the fitness function is distributed among the processes[14]. The most popular parallel models are population-based where the population itself is distributed in niches[15], also called islands sometimes[9]. Such models require a periodic migration of individuals between the sub-populations. Theshared-memory GAs are a subset of the parallel and distributed models. The parallelization techniques can be ported and adapted from one type of architecture to the other, but with specific features that can be optimized in each case. A survey of these algorithms can be found in[1].There are positive arguments in favor of models based on population division, such as a high degree of independence for each process. Among the drawbacks we can cite the fragmentation of the population into small pieces. This can generate issues such as the premature converge of the population to a local optimum or general loss of diversity. Larger populations have been reported to perform better by several studies or even to be necessary to the success ofparallel implementations[3].In a different direction, coevolutionary algorithms have interested and fascinated researchers for a good number of years. Even though competitive coevolution is the more popular form, the cooperative form has been proven to give good results[2]. These approaches decompose the problem into parts evolving separately[12]. For the purpose of the fitness evaluation, these parts are assembled into a complete chromosome.[7] argues that it is not the separability of the problem that makes these approaches successful, but their increased exploratory power. Some theoretical studies of the conditions under which these algorithms can achieve the global optimum have been proposed[10].The model that we propose in this paper bridges the gap between these two approaches. It is a variant of the cooperative coevolutionary approach designed for shared memory parallel architectures. While the usual cooperative approach is implemented for problems that are naturally divisible into subpopulations, our model generalizes this technique by making it applicable to any problem. Our model is based on a division at the genotype level of the population into several agents or processes. It is not an algorithmically equivalent version of the genetic algorithm, or of a standard cooperative coevolutionary algorithm, but a hybrid model designed for parallel architectures. This model can potentially run faster and achieve better results than the standard GA. In our approach, each process receives a partial chromosome to evolve. All the genetic operations are restricted to this subset of genes. For evaluation purposes, a template is kept by every process containing information about the best genes found by all of the other processes up to that point. A periodic exchange procedure keeps this information up to date.Finally, when aiming to optimize the genetic algorithms for massively parallel architectures, it becomes undesirable both to split up the population into too small nests, and to divide the chromosome too much. A hybrid approach can be a good compromise and for this purpose both population and chromosome division models need to be studied thoroughly. This paper contributes to the study of the less observed of the two approaches.The paper is structured the following way. Section 2 presents the details of our parallel model for the genetic algorithms. Section 3 introduces the three test problems that we used for out experiments. Section 4 shows the experimental results and the paper ends with conclusions.
2. Chromosome Division Model
- Our model for parallel genetic algorithms follows a similar idea to the one described in[16]. The difference is that the current model is implemented for shared memory architectures as opposed to a Beowulf cluster, and the experiments use a different set of problems. Preliminary results were also presented in[17,18], although the set of problems used here is almost entirely new.
2.1. Problem Division
- According to the most popular approach to parallel genetic algorithms, which is the island one, the population is decomposed in several islands or niches, each of them evolving in parallel. In such a model, the evolution in each population is self-contained, and the only thing that makes it a unified process is an occasional migration of individuals between the islands.Our motivation comes from the fact that smaller populations can more easily lead to suboptimal solutions and premature convergence. The chromosome division allows us to maintain a larger population for each process using the same amount of memory.The idea behind this parallel model is that the problem to be solved is divided into several tasks. Each task is then assigned to a different process that will focus on it while exchanging information with the other processes. This is similar to a multi-agent approach that has been proved efficient for many applications before, in a variety of contexts.Thus, in the approach proposed in this paper, the division happens along the genotype. The genes composing each chromosome are divided among the processes, such that the task of each process consists of evolving a pre-determined part of the chromosome. Each process performs the standard genetic operations on its subset of genes. When the fitness evaluation is needed, the subset of genes is inserted into a global template that allows it to be seen as a complete chromosome, as shown in Figure 1. The top chromosome in this figure represents the template, which is a chromosome with some missing genes. The pieces marked as “Population for P1” represent partial chromosomes that complete the genes missing in the template. They are assembled together into a complete chromosome that can be evaluated with the usual fitness function. This template is periodically exchanged between the processes.Our intention was to develop a model that can be applied regardless of the separability of the problem, but the test problems we chose are also designed to show how this aspect influences the fitness performance and the speedup.All the genetic operators are performed according to their standard definition, with the exception that they are restricted to the subset of the genes assigned to each process. Let n be the size of the chromosome, with the indexes for the genes going from 0 to n-1. Let us suppose that we have p processes. Each process will receive a part of the chromosome of size np = n/p. Then the process or agent with identity number id, 0 ≤ id ≤ p-1 will be in charge of the genes in the interval[id*np, (id+1)*np - 1].
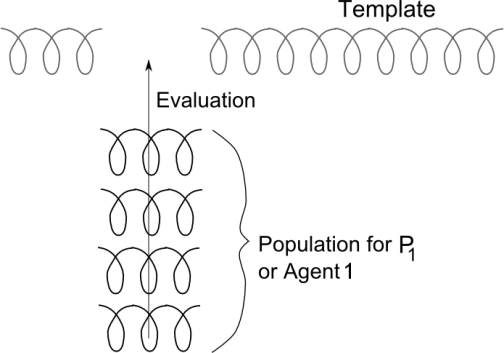 | Figure 1. A template for the population evolved by one of the agents |
2.2. Fitness Evaluation
- There are a good number of benchmark fitness functions for genetic algorithms that are separable, meaning that they can be divided into sub-problems such that the evaluation of each of them can be accomplished independently. Our model is not restricted to these types of problems specifically, but is rather designed in a general way so that it can be applied to any fitness function. However, the evaluation procedure can be optimized for separable functions and a greater performance can be achieved in terms of execution time and use of each CPU core. One of out test problems will showcase this situation.We start with the assumption that to evaluate the fitness function for any combination of genes, we need a full set spanning from 0 to n-1. Thus, to evaluate a partial chromosome, we need to complete it with the template. The evaluation consists of plugging the partial chromosome into the common template, and then passing this complete individual to the fitness function.An exchange procedure insures that the template is kept reasonably up to date with respect to the latest best performing genes obtained by each process. During the exchange phase, each process copies the genes of the best chromosome found so far in terms of fitness to a global “best chromosome” shared by all the processes. After all of the processes have finished this update, each of them updates its own template from the global best chromosome. This procedure takes place periodically and can involve a synchronization of the processes in terms of number of generations produced in between the exchanges. Since only one chromosome is exchanged in the process, our model is coarse-grained. The next section talks about this aspect more in detail.
2.3. Synchronous vs Asynchronous Exchange
- The only communication between the processes happens during the exchange procedure. From a synchronization point of view, we propose to compare two approaches. In the first one, each exchange phase happens after the exact same number of generations for each process, and this is accomplished through a barrier call. In this model, called synchronous, all the processes evolve approximately at the same time and during the exchange they need to wait for all of them to reach the entry point in order to proceed. The genes in each chromosome represent a fairly homogeneous evolutionary step during the fitness evaluation.In the second approach, called asynchronous, each process can update the global best chromosome periodically without having to wait for any of the others. Thus, genes evolved in substantially different generations are combined for the evaluation of the fitness.The synchronous exchange procedure is shown below in C++ based pseudo code. In this algorithm we assume that the indexes in the partial chromosome are kept consistent with the position of the genes in the complete chromosome. To make the procedure easier to understand, the id of the process is used as an index for the best partial chromosome and for the template. Practically, our implementation is object oriented, the exchange function is a class method, and these variables marked with the id are class attributes. The global variables shared by all the processes are shown with capitalized names.void Exchange_synch(int id) {np = Chromosome_Size/ Number_Of_Proc;Barrier(Number_Of_Proc); for (i=id*np; i<(id+1)*np; i++)Best_Chromosome[i] = best_partial_chr[id][i];Barrier(Number_Of_Proc);for (i=0; i
3. Test Problems
- We have chosen three problems to test our parallel model with, two of them being of the benchmark type, and one a real-world problem. The specifics of each of them should allow us to showcase different features of our program. The benchmark problems consist of fitness functions of linear complexity over the number of genes and also uniformly fast to compute.The real-world problem is computationally more expensive and non uniform over the set of chromosomes. For this last function, a global optimum is not known.The two benchmark problems are chosen with a fitness that is linear over the chromosome length to show the speedup potential of our model for the most common category of problems. These functions are very similar to other problems used for benchmarking in various studies. The real-world problem is a difficult one chosen to showcase the potential of our model in terms of quality of solutions.A second aspect that differentiates these problems is the reciprocal influence of genes at different locations in the chromosome in the computation of the fitness or separability. For the real-world problem, such influences are present, and a good performance cannot be achieved in the absence of proper process coordination. For the first benchmark problem, there is an even higher degree of reciprocal influence of the genes from one process to another than for the real-world problem. For the second benchmark problem the fitness influence is localized to the genes assigned to each process, meaning that this function is highly separable, and the algorithm is optimized to take advantage of it. Thus, we hope to show how our model can behave in each of these three situations.For reasonable comparison grounds for the three problems we have used the same experimental settings as much as possible: population size (50), number of generations (1000), and chromosome size (360). The number of genes is determined by the number of parameters defining the real-world problem, and we were able to configure the two benchmark problems with the same value.
3.1. Benchmark Problems
- The first problem is known in the literature as the Rosenbrock function[8,13] or the DeJong function[6], and as a difficult optimization problem. It consists of minimizing the following function:
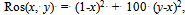 | (1) |
|
3.2. Real-Life Problem
- The third problem we are using consists of optimizing the parameters defining a pilot for a simulated motorcycle. For this problem, the evaluation requires significantly more computations, and thus it will allow us to observe the improvement in performance in that respect. Contrary to the linear functions, the complexity of evaluating a chromosome is not uniform, but can vary significantly from one individual to the next. This constitutes an additional challenge for the parallel model. This function is partially separable.The physical model of the motorcycle has been more extensively described in[19] and is close to[4]. The motorcycle is modeled as a system composed of several elements with various degrees of freedom, consisting of position and orientation on the road, speed, rotation of the handlebars, and leaning. The driver's input into the system is defined by the tuple u=(τ, βf, βr, φ, α) where τ is the acceleration in the direction of movement provided by the throttle/gear control, βf, βr are forces applied on the front and rear brakes respectively, φ is the leaning angle, and α is the handlebar turning angle. This driver can be either a human player or an autonomous agent controlling the vehicle.The movement is defined by Newtonian mechanics, where the acceleration is defined by gravity, friction, drag, and the throttle. The brakes are factored into the friction force.The autonomous pilot uses perceptual information to make decisions about the vehicle driving. This information consists in the visible front distance, the lateral distance to the border of the road from the current position of the vehicle and from a short distance ahead of the vehicle, and the slope of the road.The motorcycle is driven by several control units (CUs), each of them controlled by an independent agent. The current CUs are the gas (throttle), the brakes, and the handlebar/leaning. Each of these CUs is independently adjusted by an agent whose behavior is intended to drive the motorcycle safely in the middle of the road at a speed close to a given limit. The agents behave based on a set of equations relating the road conditions to action. The full set of equations is described in[19]. The equations comprise a fair number of coefficients and thresholds, and these are the values that are evolved by genetic algorithms.To apply the GA to this problem, we chose a representation where each configurable coefficient is assigned 10 binary genes, and the chromosome results by concatenating all of the coefficients. As we have 36 coefficients, the chromosome has a length of 360.A chromosome is evaluated by running the motorcycle in a non-graphical environment once with the pilot configured based on values obtained by decoding the chromosome over a test circuit presenting various turning and slope challenges. Each run can end either by completing the circuit, or by a failure condition. A failed circuit can be caused by one of the following three situations: a crash due to a high leaning angle, an exit from the road with no immediate recovery, or crossing the starting line without having reached all the marks, as when the vehicle takes a turn of 180 degrees and continues backward.To compute the fitness we marked 50 reference points on the road and counted how many of them were reached by the motorcycle with a given degree of approximation. The fitness is computed as follows:
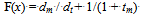 | (2) |
4. Experimental Results
- In this section we present some of the experimental results with our model testing both the executions time/speedup and the fitness performance. Table 2 introduces the platforms that we have used for our computations. In the operating system column, XP stands for Microsoft Windows XP, W7 stands for Microsoft Windows 7, OsX stands for the Mac OsX 10.5.6, and Ub stands for Ubuntu 8.04. The program was implemented in standard C++ using the pthread library and its local version for each platform. The code that is being run is the same on all of the platforms for each reported experiment.For the experiments concerning the speedup, there is no point in comparing approaches that don't perform a comparable number of computations. Since the speedup is our foremost interest here, we have chosen to run all the experiments with the same number of generations.
4.1. Synchronous versus Asynchronous Exchange
- The first set of experiments compares the synchronous versus asynchronous exchange models on different platforms for a variety of number of processes.Table 3 shows the average execution time as number of seconds for the Rosenbrock function. Table 4 shows the average execution time for the deceptive problem. The chromosome length is 360, the population is of size 50, and we have run 1000 generations in all the cases. The results are averaged over 100 runs. The column labelled “Com” identifies the exchange function as synchronous (S) or asynchronous (A).
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||
4.2. Influence of the Synchronization Period
- The second set of experiments focuses on the aspect of the number of generations between the synchronization and exchange phases and on how it influences the overall performance. For this purpose we chose the Rosenbrock function because it presents the highest degree of dependence of the genes on each other for the fitness.We have run these experiments with a synchronization period taking several values from 1 to 1000. For this set of experiments we have used the Mac OsX platform with the Core 2 Duo processor.Figure 2 shows the speedup obtained under various settings of the parameter, where the legend indicates the number of processes and the synchronization type (S for synchronous, A for asynchronous). The speedup improves a substantial amount when going from a period of 1 to one of 5, and from 5 to 10, but after that the improvement slows down. It is also interesting to note that for a high amount of synchronization, the synchronous model is faster than the asynchronous model for every value of the number of processes, but with less synchronization, the asynchronous model eventually becomes faster.
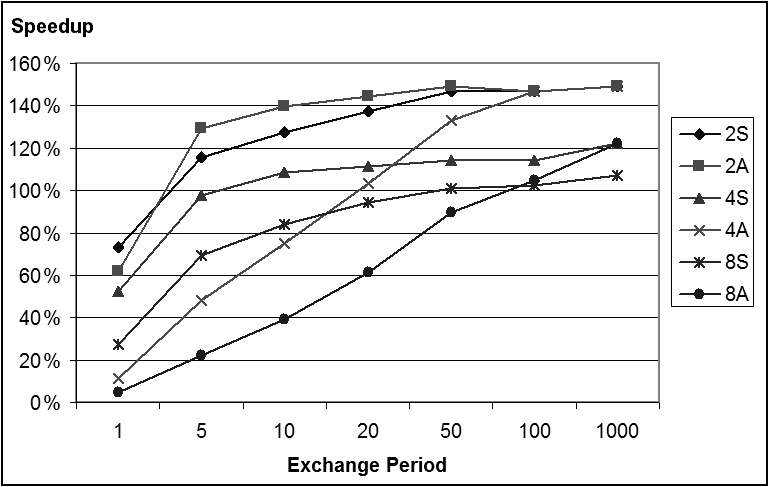 | Figure 2. Speedup for the Rosenbrock function as a function of the exchange period |
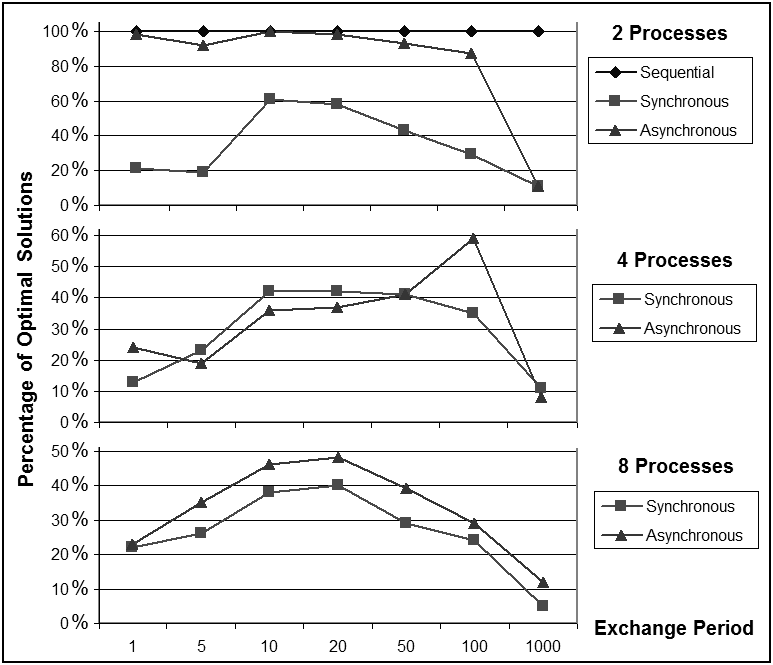 | Figure 3. Percentage of optimal solutions found as a function of the exchange period |
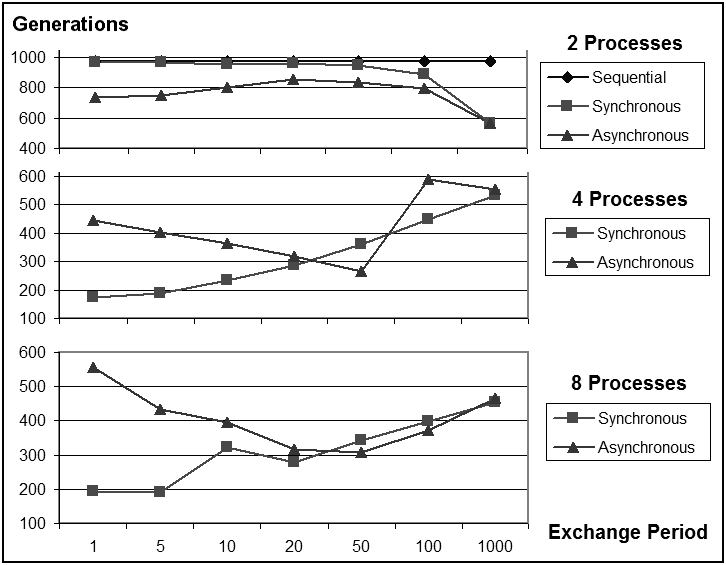 | Figure 4. The number of generations to achieve the fitness as a function of the exchange period |
| ||||||||||||||||||||||||||||||||
5. Conclusions
- In this paper we presented a shared memory parallel model of genetic algorithms designed to take advantage of multiple CPU cores in common current architectures. We have tested our model with three sets of problems of various difficulties on four different platforms with several types of processors and operating systems. Each problem presents different separability properties.The experimental results presented in Section 4 explore the performance of the parallel model on several levels. First, in what concerns the speedup, for the benchmark functions there is a clear improvement on the platforms with multiple CPUs. A more modest but still noticeable speedup can be observed as well for the more difficult problem of configuring the autonomous pilot. The best speedup is around 185% on 2 CPU cores and around 223% on 4 CPU cores.In terms of average fitness achieved in 1000 generations, for all test problems we can observe that the parallel model outperforms the sequential model for a number of processes less or equal to 4, which is also the maximum number of available cores on our test platforms. For the Rosenbrock function, the sequential model was able to find the optimal solution 100% of the time in 1000 generations, and this is also the case for the asynchronous model with 2 processes. For the deceptive problems, we see about 12% fitness improvement in the best case. For this problem, the parallel model was able to fine-tune the search to smaller parts of the chromosome and thus, improve the performance. For the motorcycle problem, we see an improvement of 3.5% in the best case.A comparison of the synchronized and asynchronized schemes shows an improvement in speedup for the asynchronous model without loss in performance. Another set of experiments have shown that the synchronization and communication period of 10 generations that we have chosen is a nearly optimal choice of balancing between the speedup and fitness performance for both the synchronized and asynchronous models.On the subject of problem separability, several conclusions can be drawn from our experiments. The speedup can be better improved for separable problems, which is to be expected. The fitness improvement is also more impressive for the more separable problems. A higher division of the chromosome is also more beneficial to problems that are more separable. With a division into 2 processes, though, an improvement can be observed for all the problems, even the non-separable ones. This means that even though our algorithm is more efficient for separable problems, it can still present some advantages even for non-separable ones. In conclusion, our model presents a valid approach to taking advantage of the multi-core computing technologies that are now widely available, even for non-separable problems, and a strict synchronization between the processes is not a benefit in terms of performance.
References
| [1] | G. Dozier, “A comparison of static and adaptive replacement strategies for distributed steady-state evolutionary path planning in non-stationary environments”, International Journal of Knowledge-Based Intelligent Engineering Systems (KES), vol. 7, no. 1, pp. 1-8, January 2003. |
| [2] | A.-M. Farahmand, M. N. Ahmadabadi, C. Lucas, and B. N. Araabi, “Interaction of culture-based learning and cooperative co-evolution and its application to automatic behavior-based system design”, IEEE Transactions on Evolutionary Computation, vol. 14, no. 1, pp. 23-57, 2010. |
| [3] | F. Fernandez, M. Tomassini, and L. Vanneschi, “An empirical study of multipopulation genetic programming”, Genetic Programming and Evolvable Machines, vol. 4, no. 1, pp. 21-51, March 2003. |
| [4] | N. Getz, “Control of balance for a nonlinear nonholonomic no-minimum phase model of a bicycle”, in American Control Conference, Baltimore, June 1994. |
| [5] | D. E. Goldberg, K. Deb, and J. Horn, “Massive multimodality, deception and genetic algorithms”, in R. Manner and B. Manderick, editors, Proceedings of Parallel Problem Solving from Nature II, pp. 37-46, 1992. |
| [6] | R. Irizarry. Lares, “An artificial chemical process approach for optimization”, Evolutionary Computation, vol. 12, no. 4, pp. 435-459, 2004. |
| [7] | T. Jansen and R. P. Wiegand, “The cooperative coevolutionary (1+1) EA”, Evolutionary Computation, vol. 12, no. 4, pp. 405-434, 2004. |
| [8] | S. Kok and C. Sandrock, “Locating and characterizing the stationary points of the extended Rosenbrock function no access”, Evolutionary Computation, vol. 17, no. 3, pp. 437-453, 2009. |
| [9] | T. Van Luong, N. Melab, and E. Talbi, “GPU-based island model for evolutionary algorithms”, in Proceeding of the Genetic and Evolutionary Computation Conference, pp. 1089-1096, Portland, OR, July 7-11 2010. |
| [10] | L. Panait, “Theoretical convergence guarantees for cooperative coevolutionary algorithms”, Evolutionary Computation, vol. 18, no. 4, pp. 581-615, 2010. |
| [11] | O. Parinov, “The implementation and improvements of genetic algorithm for job-shop scheduling problems”, in Proceedings of the Genetic and Evolutionary Computation Conference, pp. 2055-2057, Portland, OR, July 7-11 2010. |
| [12] | M. Potter and K. De Jong, “Cooperative coevolution: an architecture for evolving coadapted subcomponents”, Evolutionary computation, vol. 8, no. 1, pp. 1-29, 2000. |
| [13] | H. H. Rosenbrock, “An automatic method for finding the greatest or least value of a function”, The Computer Journal, no. 3, pp. 175-184, 1960. |
| [14] | Y. Sato, M. Namiki, and M. Sato, “Acceleration of genetic algorithms for Sudoku solution on many-core processors”, in Proceeding of the Genetic and Evolutionary Computation Conference, pp. 407-414, Dublin, IrelandOR, 2011. |
| [15] | I. Sekaj and M. Oravec, “Selected population characteristics of fine-grained parallel genetic algorithms withre-initialization”, in GEC '09: Proceedings of the first ACM/SIGEVO Summit on Genetic and Evolutionary Computation, pp. 945-948, New York, NY, USA, 2009. |
| [16] | D. Vrajitoru, “Parallel genetic algorithms based on coevolution”, in R.K. Belew and H. Juillé, editors, Proceedings of the Genetic and Evolutionary Computation Conference, Late breaking papers, pp. 450-457, 2001. |
| [17] | D. Vrajitoru, “Asynchronous multi-threaded model for genetic algorithms”, in Proceedings of the Midwest Artificial Intelligence and Cognitive Science Conference (MAICS), pp. 44-50, South Bend, IN, April 17-18 2010. |
| [18] | D. Vrajitoru, “Shared memory genetic algorithms in a multi-agent context”, in Proceedings of the Genetic and Evolutionary Computation Conference (GECCO) (SIGEVO), pp. 1097-1104, Portland, OR, July 7-12 2010. |
| [19] | D. Vrajitoru and R. Mehler, “Multi-agent autonomous pilot for single-track vehicles”, in Proceedings of the IASTED Conference on Modeling and Simulation, Oranjestad, Aruba, 2005. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML