-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Biomedical Engineering
p-ISSN: 2163-1050 e-ISSN: 2163-1077
2022; 11(1): 1-12
doi:10.5923/j.ajbe.20221101.01
Received: Mar. 8, 2022; Accepted: Mar. 21, 2022; Published: Apr. 22, 2022

Assessment of a Neural Network Based on Texture Features Analysis: The Impact of Classifying Cancer Types Using Image Processing
Ihab S. Atta 1, 2, Ashraf S. Emam 3, Ali H. Al-Boghdady 4, Saeed M. Badran 5, Mohamed F. El-Refaei 6, Ossama B. Abouelatta 7
1Department of Pathology, Faculty of Medicine, Al-Baha University, Al-Baha, KSA
2Department of Pathology, Faculty of Medicine, Al-Azhar University, Cairo, Egypt
3Department of Automotive and Tractors Engineering, Faculty of Engineering, Mataria, Helwan University, Cairo, Egypt
4Department of Mechanical Engineering, Faculty of Engineering, Al-Baha University, Al-Baha, KSA
5Department of Electrical Engineering, Faculty of Engineering, Al-Baha University, Al-Baha, KSA
6Department of Biochemistry, Faculty of Medicine, Al-Baha University, Al-Baha, KSA
7Department of Production Engineering & Mechanical Design, Faculty of Engineering, Mansoura University, Mansoura, Egypt
Correspondence to: Ossama B. Abouelatta , Department of Production Engineering & Mechanical Design, Faculty of Engineering, Mansoura University, Mansoura, Egypt.
| Email: |  |
Copyright © 2022 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Traditional histopathology examination remains a serious task in cancer identification and is clinically vital to division the cancer tissues and group them into numerous classes. However, the diagnostic process is subjective, and the variations among technical observers and time consumed are considerable. Reliable, automated cancer detection assistance is currently an increasingly important task in the medical field. This study aims to classify different cancer tumor types. A comprehensive analysis of a new classification technique based on image processing and composition properties was performed. A graphical user interface (GUI) program dedicated to the classification and identification of cancer cell images was developed and created in-house using Matlab package. As a result, the data can improve the diagnostic capabilities of physicians and reduce the time required for precise diagnosis. The average discrimination rate demonstrates the validity of the proposed technique in distinguishing between benign and malignant lesions. This simple procedure is an encouraging application of digital image processing performance in the histopathology field compared with traditional methods. Further investigations in the future may demonstrate a great advantage in the prediction and classification of cell morphology and cancer grading using the computed segmentation technique.
Keywords: Cancer, Histopathology, Automatic classification, Image processing, Texture features, Neural network
Cite this paper: Ihab S. Atta , Ashraf S. Emam , Ali H. Al-Boghdady , Saeed M. Badran , Mohamed F. El-Refaei , Ossama B. Abouelatta , Assessment of a Neural Network Based on Texture Features Analysis: The Impact of Classifying Cancer Types Using Image Processing, American Journal of Biomedical Engineering, Vol. 11 No. 1, 2022, pp. 1-12. doi: 10.5923/j.ajbe.20221101.01.
Article Outline
1. Introduction
- Cancer is considered a major cause of death today. Despite recent advances in flexibility and awareness, cancer is not yet cured. Different types of cells cause different types of cancer [1]. In all types of cancer, the cells are abnormal and tend to populate at an extremely fast rate. Cancer cells that continue to multiply and form a lump or growth of tissue are called malignant tumors. Malignant tumors cause damage by invading tissues and organs. Moreover, inventors estimated that there will be sixteen million unique cancer cases within the next upcoming five years [2]. It is therefore essential that new therapeutic options are generated in addition to the current main management modalities of surgery, immunotherapy, and radiotherapy [3-5].When cancer is detected, the treatment is more effective if the disease is diagnosed at an early stage. The common and traditional method of cancer diagnosis is a microscopic analysis of small biopsy samples. In such an examination, histopathologists analyze the biopsy samples using microscopy and diagnose the tissue as benign or malignant based on the morphology of the tissues. Benign and malignant tissues differ substantially in their morphology [6].A morphological study is done by examining the tissue selected by a light microscope. However additional features of tissue examined may be evoked using an electron microscope. Furthermore, the study of histologic images is of great interest for both clinical diagnosis and notifications regarding both prognostic and therapeutic purposes [7,8]. The histological examination of biopsies to identify and unify disease has a critical role in both the clinical part and elementary feeding of knowledge [9]. For biopsy obtained for cancer diagnosis, the pathologist has to visualize the uniformity of cell morphology as well as tissue spread, identify the cellular changes in the level of malignancy, and determine its degree according to the rules applied. Such these studies have been incriminated to a large extent to identify malignancy including prostatic tissue [10,11], both female and male breast [12-14], uterine cervix [15,16], pulmonary tissues [17] cancer categorizations; as neuroblastoma [18]; and follicular lymphoma and its grading [19].The diagnostic process is also affected by diagnostic variance, which can be critical and necessary; moreover, the process is time-consuming and labor-intensive. Also, it is important to segment the regions of interest drawn by experts using manual methods.Recently, biopsy images grouping has become an active field of research and by using microscopic analysis, cancer can be diagnosed. The process is somewhat subjective and may lead to different inter-and intra-observer variations, which increases the demand for computer-aided cancer detection [20]. A critical task in cancer diagnosis is labeling histopathology images as having regions that are cancerous or not. Cancer tissues also need to be segmented as clinically important [21]. The most significant impact of recent research is the current dissemination of a useful and efficient automatic grading system for hepatocellular carcinoma biopsy images [22], benign and malignant mass types in mammograms [23], and ischemic stroke [24], which provides several advanced methods for image preprocessing, feature extraction and image classification. In addition, automated detection, discrimination and classification of benign and malignant features is presented in skin lesions, pigmentation [25], leukocytes [26], ultrasound liver [27] and granularity of melanoma [28].Cancer is still recognized by the visual attention of imaging studies in a qualitative model. However, human visual grading for biopsy images is very time-consuming, subjective, and inconsistent due to observer variation. Analyzing biopsy images requires a more reliable and reproducible path. Therefore, this research develops an automatic system that involves building a computer program based on a combination of two methods, namely, texture features and neural networks, to classify images of cancer cells.
2. Related Work
- Recently, it has become more common to use computers to examine, explore, and interpolate images. Related work can be grouped into three main categories: histopathological image classification methods, texture features, computer vision, and artificial neural networks.
2.1. Gray-Level Co-occurrence Matrix and Texture Features
- Texture can be considered as a visual pattern with a uniform appearance that is not affected by a single color such as water and clouds [29]. One of the most popular texturing techniques is the comparison of second-order statistics computed from stored images and queries. Measures of image texture, such as directionality, parallax, regularity and roughness [30] or degree of directionality, periodicity and randomness [31], can be computed from these images. Alternative methods of texture analysis for retrieval include the use of Gabor filters [32] and fractals [33]. Image transformation, feature extraction, texture synthesis, pattern classification, texture shape and image segmentation are the visual impressions of visual texture [34]. Haralick proposed the co-occurrence matrix, the most popular second-order statistical feature among texture features [35]. The two steps proposed by Haralick for feature extraction are: (1) computing the co-occurrence matrix and (2) computing the texture features based on the co-occurrence matrix. The proposed techniques are used in many different fields of image analysis, from remote sensing to biomedical applications. The gray level co-occurrence matrix (GLCM) is defined as follows [35]:
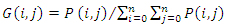 | (1) |
2.2. Histopathology Image and Texture Features
- Regarding the segmentation of histopathological images, the proposed multi-instance clustering learning process performs medical image classification, segmentation and patch-level (malignant and benign) clustering [21]. Additionally, Rathore et al. [20] proposed a colon biopsy image classification system based on statistical moments of color components and Haralick features. In addition, two automated methods of Rouhi et al. [23] are used for mammogram diagnoses. Furthermore, Cavalcanti and Scharcanski [25] developed a new method to classify skin lesions based on texture features. An overview of image exploration methods in the field of histopathology for automated cancer detection and sorting was recently presented [37]. Stock et al. [28] analyzed and differentiated the granularity of melanoma from similar areas of non-melanoma skin lesions.
2.3. Neural Networks
- An extremely complex system that can solve highly complex problems is the human brain, Figure 1(a). The most important building blocks of the brain are neurons, which are made up of many different elements. Neural networks receive impulses from all five senses and send them to muscles to perform movements or speak. A single neuron can be viewed as an input/output machine, waiting for spikes from neighboring neurons and sending spikes to other neurons when they receive enough spikes [38].Artificial neurons behave like biological neurons, Figure 1(b). An artificial neuron has input connections that are assembled to find the strength of its output, which is the result of the entire feed activation function. There are several activation functions, the most common is the sigmoid activation function, which returns a number between 0 and 1, where 0 is the low input value and 1 is the high input value. The function then takes the result and passes it as input to other neurons through other individually weighted connections. These weights then show the network behavior. Human brain connects neurons in seemingly random order, then fires asynchronously.
 | Figure 1. Basic knowledge of ANN |
3. Material and Methods
- One hundred samples were taken from different tissues, including colon, prostate, testis, skin, and uterine tissue. A total of 20 different tissue types were collected, in which benign and malignant lesions were evenly distributed. All selected tissues were diagnosed, so we divided them into two broad categories: lesions without atypical or malignant features (10 cases) and lesions with malignant features (10 cases). All cases are conducted in accordance with relevant guidelines and regulations. This study was conducted under No. 29/2021 with the approval of the relevant ethics committee of the Faculty of Medicine, Al Baha University. This approval includes acceptance of all research methods. In addition, all participants in this study obtained written consent, which included consent to publish.
3.1. Specimen Collection and Preparation
- All specimens used in this study were collected from archival materials in the Department of Pathology at KSA. For histopathological examination, all cases were routinely fixed with 10% formalin, embedded in paraffin, sectioned at a thickness of 4 μm, and stained with hematoxylin and eosin.
3.2. Histopathological Examination
- To confirm the diagnosis, two pathologists inspected all slides. In two confusing cases of prostate lesions that have features overlapping with malignancy, these cases were stained immunohistochemically by high-molecular-weight cytokeratin (HMWCK) (clone 34βE12) antibodies to obtain an accurate diagnosis.
3.3. High Molecular Weight Cytokeratin Immunostaining
- For HMWCK immunostaining, all immunohistochemical analyses were performed on routinely processed, formalin-fixed, paraffin-embedded tissue. Tissue sections were cut at 4 µm and mounted on poly-L-lysine-coated slides. Immunostaining was performed in paraffin-embedded cell lines using the HMWCK (clone 34βE12) antibody. The 4-μm sections were deparaffinized, rehydrated, and subjected to microwaving in 10 mmol/L citrate buffer, pH 6.0 (BioGenex, San Ramon, CA), in a 750 Woven for 15 minutes. Slides were allowed to cool at room temperature for 30 minutes. The 4A4 antibody (1:50 dilution) was applied at room temperature for 2 hours in an automated stainer (Optimax Plus 2.0 bc; BioGenex, San Ramon, CA). The detection steps were performed by the instrument using the MultiLink-HRP kit (BioGenex). Peroxidase activity was localized using 3,3-diaminobenzidine or 3,3-diaminobenzidine-nickel chloride. Standardized development periods allowed an accurate comparison of all samples.
3.4. Proposed System
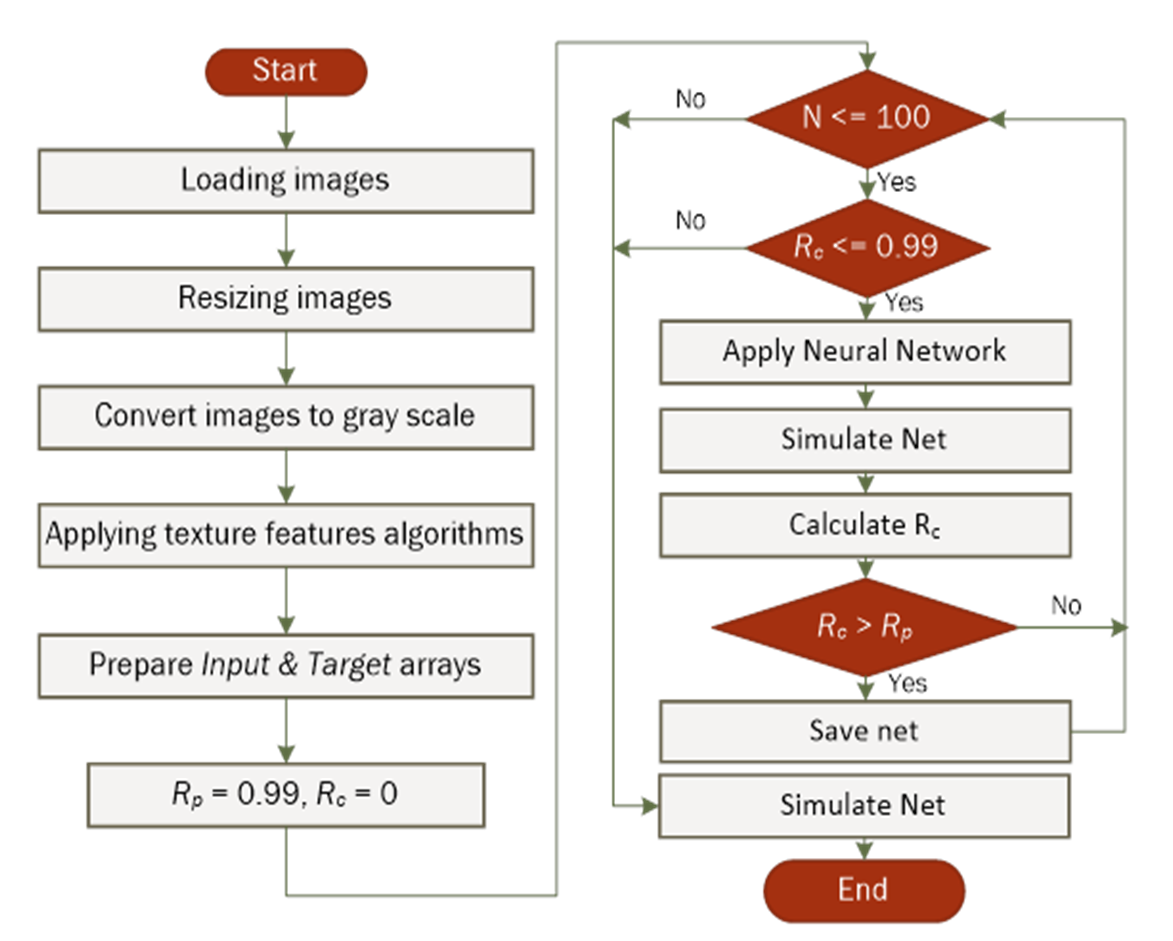
- A collection of colon, prostate, testicular, dermal, and uterine images was used in this work (n = 20 for each). The flowchart shown in Figure 2, illustrates the main steps applied during the analysis procedure.
 | Figure 2. Block diagram of the procedure |
|
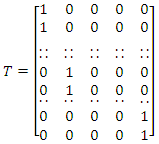 | (2) |
3.5. Automatic Classification Program
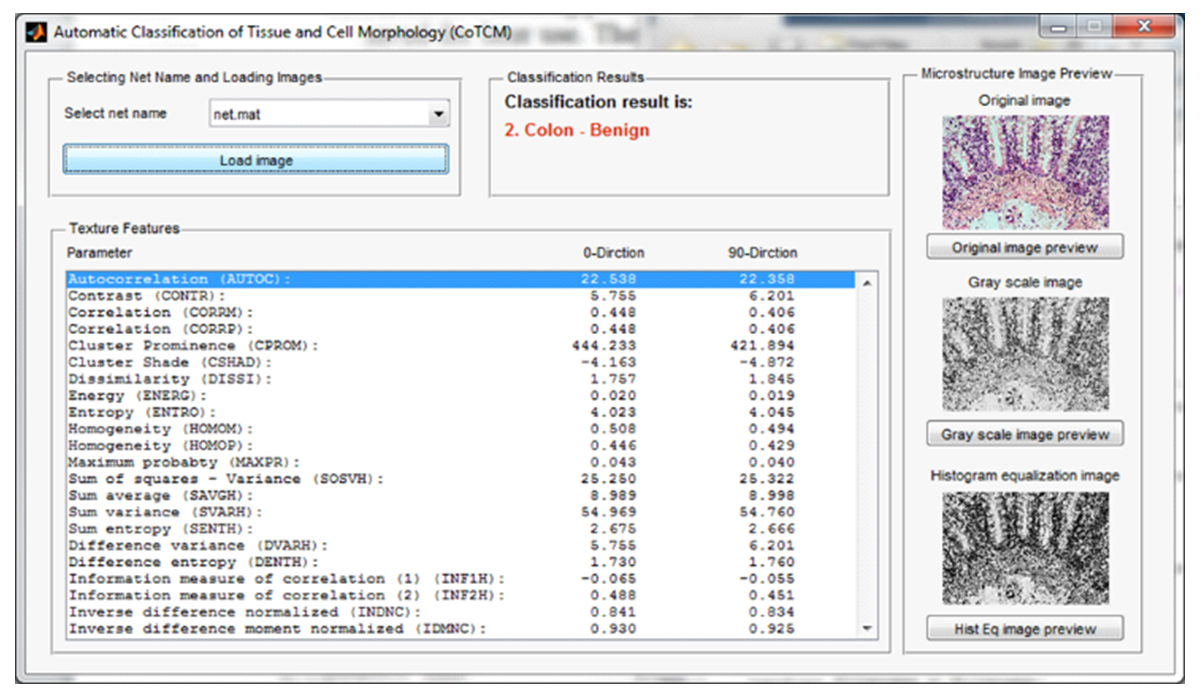
- For the classification of tissue morphology, a program named Automatic Classification of Tissue Morphology Program (ACoTM) was designed and constructed in-house using Matlab Packages [40]. Figure 3 shows the main interface of this program. The user can choose a neural network through the drop-down menu (Selecting Net Name), the image is loaded to be automatically classified, and the results of classification were displayed through the (Classification Results) area. In addition, the values of texture features are presented in the lower-left of the program window, and the original image, gray-scale images, and image after applying the histogram equalization were displayed in the right part of the program window.
 | Figure 3. The main interface of Automatic Classification of Tissue Morphology, ACoTM |
4. Results
4.1. Histopathological Examination
- The results of this study showed that all the benign lesions studied were frankly benign, with no suspicion of malignancy or even features of atypia. Initially, all cases of malignancy exhibited features of the malignant criteria, namely hyperchromasia, increased nucleocytoplasmic ratio, prominent nucleoli, and pleomorphism.
4.1.1. Colonic Specimens
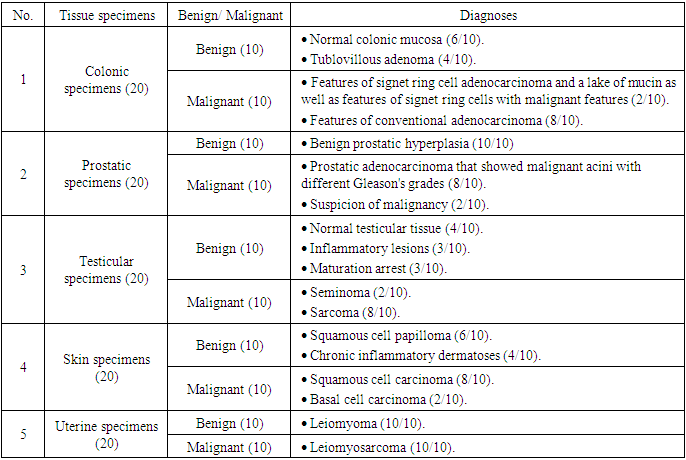
- Ten colonic specimens showed six cases of normal colonic mucosa (60%) and four cases of tubulovillous adenoma (40%). Ten cases of colonic carcinoma contained two cases (20%) with features of signet ring cell adenocarcinoma and a lake of mucin as well as features of signet ring cells with malignant features and eight cases (80%) with features of conventional adenocarcinoma, Figure 4.
4.1.2. Prostate Specimens
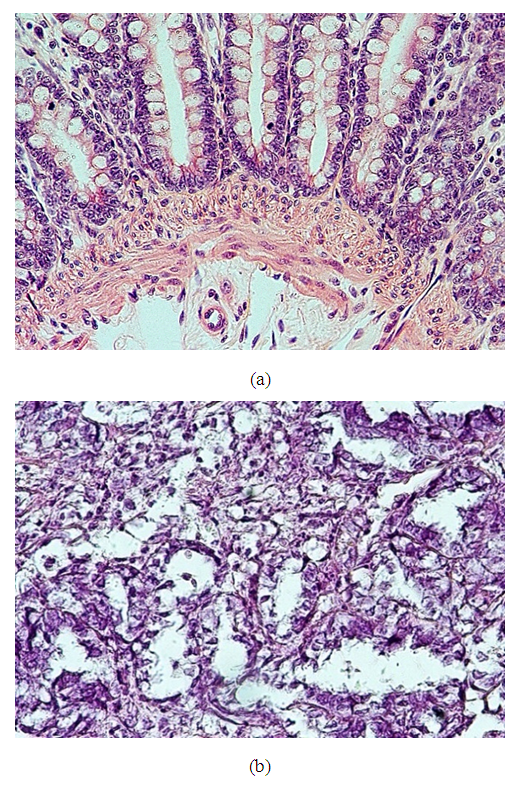
- Twenty prostate specimens revealed 10 cases of benign prostate hyperplasia and eight cases of prostate adenocarcinoma that showed malignant acini with different Gleason's grades. Two cases with suspicion of malignancy underwent immunohistochemical study by staining with an HMWCK (clone 34βE12) antibody to highlight the basal layer of acini. Cases with high positivity staining were considered to be benign lesions, and the negative cases were considered to be malignant lesions as the absence of a basal cell layer is a cut point for the diagnosis of adenocarcinoma. Of these cases, one case showed a retained basal cell layer, so it was identified as benign hyperplasia, and the other case revealed the absence of the basal cell layer and was identified as prostate adenocarcinoma, Figure 5.
4.1.3. Testicular Specimens
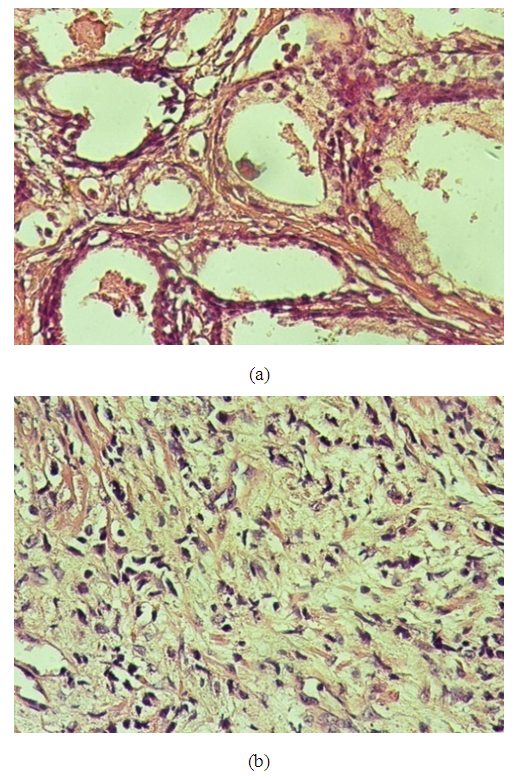
- Ten cases showed benign features: four cases were normal testicular tissue, three cases with inflammatory lesions with no evidence of malignancy and showed normal architecture and different stages of spermatogenesis, and three cases showed maturation arrest at the level of spermatid. Ten cases revealed features of malignancy: two cases (20%) were diagnosed as seminoma, and eight cases (80%) were diagnosed as sarcoma, Figure 6.
4.1.4. Dermal Specimens
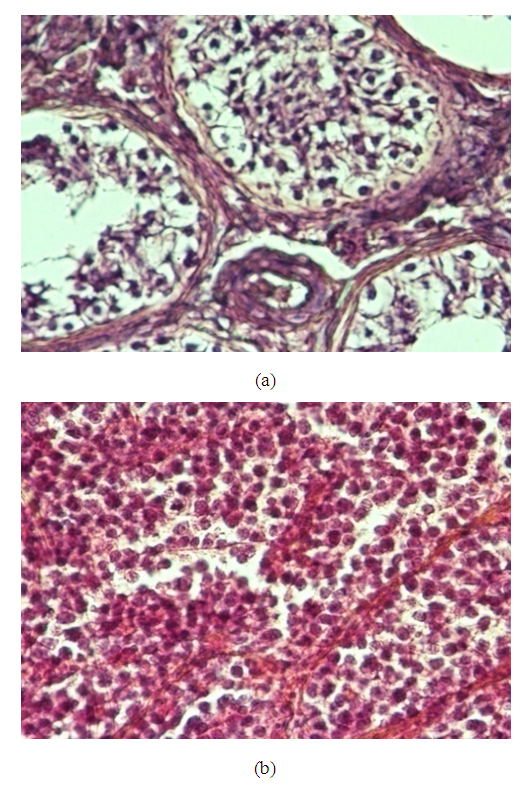
- Twenty cases of dermal specimens revealed six cases of squamous cell papilloma, four cases of chronic inflammatory dermatosis, eight cases with features of squamous cell carcinoma, and two cases with features of basal cell carcinoma. The squamous cell carcinoma cases showed features of malignant criteria in the form of squamous nests that recapitulate layers of skin with malignant criteria. Of these cases, three of eight cases (38%) were grade I, three cases (38%) were grade II, and two cases (24%) were grade III, Figure 7.
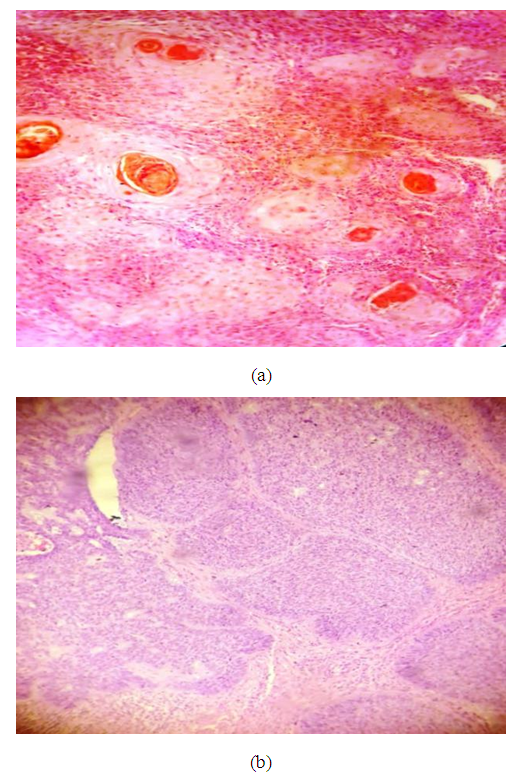 | Figure 7. (a): A case of skin biopsy showed features of squamous cell carcinoma (×200), (b): A case of basal cell carcinoma showed basaloid features with palisading towards the periphery (×400) |
4.1.5. Uterine Specimens
- Twenty cases of smooth muscles showed 10 cases of leiomyoma and 10 cases of malignant lesions. Of the malignant lesions, six cases were leiomyosarcoma and four cases were diagnosed as uterus cell sarcoma, Figure 8.
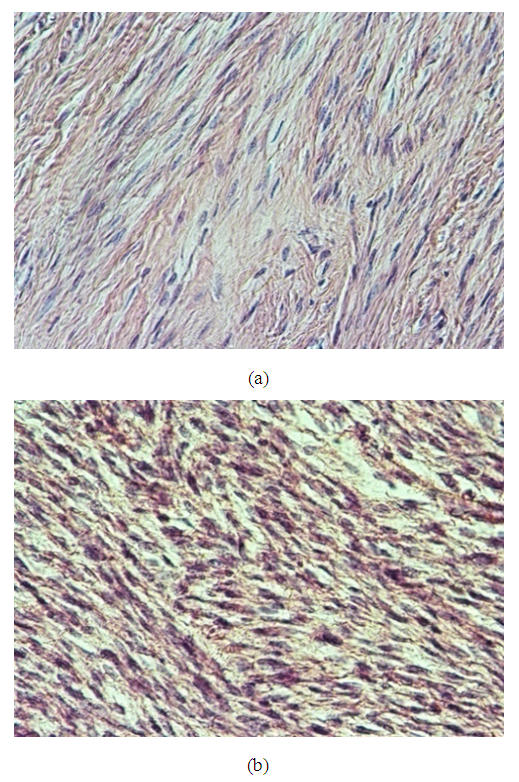 | Figure 8. (a): A case of skin biopsy showed features of squamous cell carcinoma (×200), (b): A case of basal cell carcinoma showed basaloid features with palisading towards the periphery (×400) |
4.2. Texture Feature Analysis
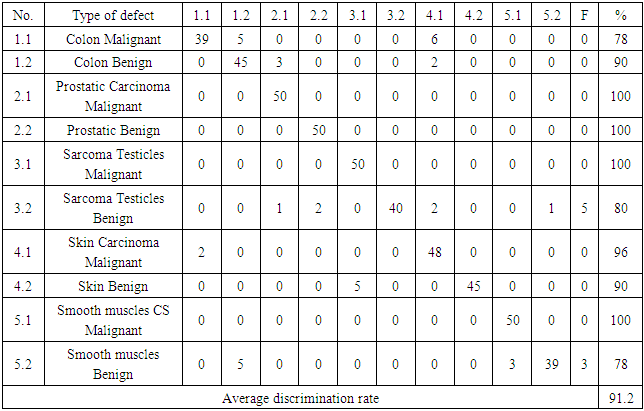
- Table 2 shows the success rates of the Automatic Classification of Tissue Morphology Program (ACoTM) in the classification of different types of cancer and classifying benign and malignant cells. The success rate is calculated as the percentage of the number of cases or images successfully classified by the program divided by the total number of images. The overall percentage discrimination rate shows that the designed program effectively achieves its purpose. The success rate of classifying different types of cancer varies from 78% to 100%.
|
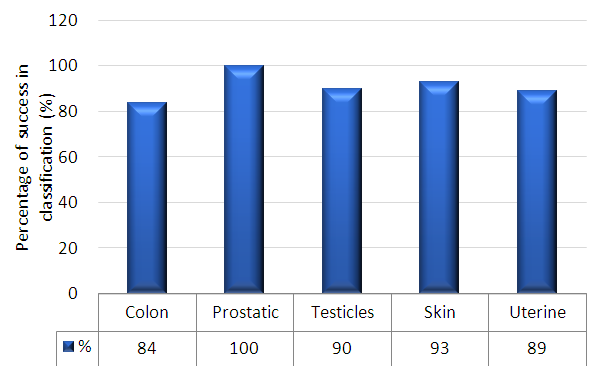 | Figure 9. Percentage success in classification for colon, prostate, testicles, skin, and uterine |
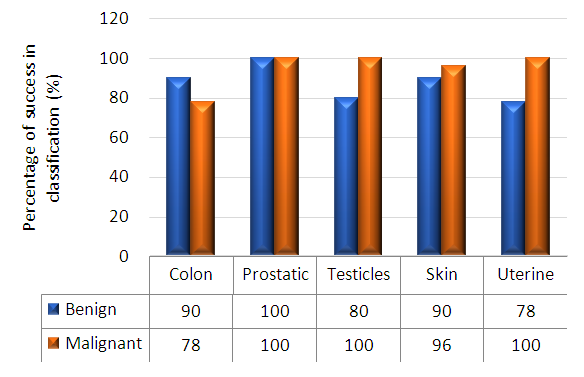 | Figure 10. Percentage success in classification for benign and malignant colon, prostate, testicles, skin, and uterine |
|
|
5. Discussion
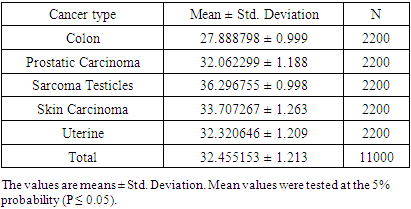
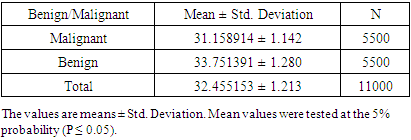
- The histopathology exploration remains a thoughtful task in cancer identification and clinically vital to division the cancer tissues and group them into various classes. Moreover, an accurate histopathology diagnosis is essential to ensure the most effective treatment for the diagnosis of cancer types. However, the diagnostic process is subjective, the technical observer is prone to inter-and intra-observer variations, and the time consumed is considerable. Therefore, a qualitative assessment of the images is essential for objective diagnoses, this shed the consideration of investigators and scientists to ensure the perspective of histopathology discrimination. Studies conducted earlier by Xue et al. [47], for histopathology image segmentation proposed that the MCIL method concurrently performs image classifications for non-cancer vs. cancer images, medical image segmentation, non-cancer vs. cancer tissues, and patch clustering, which are different classes [21]. Most of the attention over the past few years has been directed toward an automatic system for grading hepatocellular carcinoma biopsy images, which was proposed by Huang et al. to classify benign and malignant tissues [22]. Furthermore, Rajini and Bhavani presented a new approach for the automatic detection of ischemic stroke using segmentation, mid-line shift, and image feature characteristics, which separate the ischemic stroke area from healthy tissues in computed tomography images [24]. Additionally, Wang et al.’s [40] findings aimed to develop a computer-assisted algorithm for tumor segmentation and characterization using both kinetic information and the morphological features of 3-D breast DCE-MRI. A new multimodality images registration and deformation method, based on the Thin Plate Splines-Robust Point Matching (TPS-RPM) algorithm, is introduced by Makni et al. [41] to facilitate image-guided cancer ablation techniques.Furthermore, Putzu et al. [42] proposed an innovative method for the automatic identification and classification of leucocytes using microscopic images, providing an automated procedure to support the recognition of acute lymphoblastic leukemia. Their results indicated that the proposed method can efficiently identify the white blood cells present in an image and properly classify leukoblasts with great accuracy.In the present study, the histological assessment of human tissue has emerged as the key challenge for the detection of different cancers originating from different tissues, such as the colon, prostate, testicles, skin, and uterus. To gain more insights into the investigation and classification of these cancer types, we exhaustively analyze a new computed classification technique based on image processing and texture features. As a result, in this study, 100 benign and malignant specimens (50 for each) were used. Taking into account image light intensity and contrast, the number of images was increased by five times. These specimens were collected from the colon, prostate, testicles, skin, and uterus. In the case of the colon, the percent of successful classification is 78% for malignant and 90% for benign cases, and the total successful classification of colon cases is 84%. This means that the classification failed in 16 of 100 images, Figure 10. This outcome is unique for the beneficial characteristics of computed classification This allows us to distinguish between different types of cancer and cancer cases (benign and malignant). Therefore, different cancer types and cases can be classified by relying on the Automatic Classification of Tissue Morphology Program (ACoTM). It also agrees with other findings from Rathore et al. [20], which suggests that the proposed colon biopsy image classification system benefits from discriminatory capabilities of information-rich hybrid feature spaces (namely, color component-based statistical moments and Haralick features), and the performance enhancement is based on the ensemble classification methodology.Moreover, in the prostate, the percent of successful classification is 100% for both benign and malignant cases; therefore, the total successful classification of prostate cases is 100%. This means that classification succeeded in all specimen images, Figure 10. This suggestion is consistent with the work of Chen et al. [43], which presented an automatic segmentation cost function algorithm based on deformable organ models built from segmented training data of prostate and rectal tissue from 3D computed tomography. Our data are in agreement with DiFranco et al. [44], who proposed methodologies that incorporate ensemble learning for feature selections and classifications on expert-annotated images.Based on the findings of this study, our results succeeded in classifying testicular cases at a rate of 100% for malignant cases and 80% for benign cases, Figure 10. No data have been published and made available in this area using the texture feature. Adesanya et al. [45] is the only group for which an ultrasound evaluation had an accuracy of 86.5% in preoperative localization of undescended testes compared with clinical examination.In the current study, our data for the skin cases showed that the image classification results were successful at a rate of 96% for malignant cases and 90% for benign cases. This means that the classification failed in four of 100 images. This is in agreement with the results reported by Cavalcanti and Scharcanski [25], who presented methods for classifying lesions in pigmented skin as benign or malignant by using a new technique to improve the processing and analysis of skin images. Consistent with Mete et al. [46], this system introduced dermoscopy images of border-driven density-based frameworks to identify skin lesion(s). The proposed algorithm expands the regions at the borders of a cluster that then speeds up the process without losing precision or recall, unlike the conventional density-based clustering algorithms.Finally, our study demonstrates that the classification of uterine cases has a 100% success rate for malignant and a 78% success rate for benign cases. These data agree with the study demonstrated by Xue et al. [47], which reported efforts to analyze and extract significant areas from uterine cervix images in a large database created for the study of cervical cancer. By proposing new algorithms, they focused on developing open-source tools, which are synchronous with the research objectives. Thus, we can conclude that direct access to advanced diagnostic technology and clinical expertise improves the accuracy of a diagnosis to 91.2%. Advanced technology can help pinpoint tumors and allow doctors to determine the best combination of treatments in the future.
6. Conclusions
- Computational pathology is a novel field comprising aspects of machine learning, computer vision, clinical statistics, and general pathology. The principal focus of this study is to define this new field and develop and investigate statistical methods, which can be combined within a unified framework to answer scientific and clinical questions in pathology. Moreover, the development from classical image processing to statistical pattern recognition in the domain of computational pathology is marvelous. This methodology is successfully employed for several diagnostic tests, which may in the future identify an effective treatment approach from the beginning before treatment even starts. In addition, the morphological and histological feature assessment by neural networks can support cancer diagnosis and can precisely recognize cancer cell types.
ACKNOWLEDGEMENTS
- This research is a part of a project entitled "Automatic Classification of Tissue and Cell Morphology using Image Processing and Neural Network". This project was funded by the Deanship of Scientific Research, Al-Baha University, KSA (Grant No. 1435/029). The assistance of the deanship is gratefully acknowledged.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML