-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Biomedical Engineering
p-ISSN: 2163-1050 e-ISSN: 2163-1077
2011; 1(1): 20-25
doi: 10.5923/j.ajbe.20110101.04
Feature Selection and Classification of Breast Cancer on Dynamic Magnetic Resonance Imaging Using ANN and SVM
F. Keyvanfard 1, M. A. Shoorehdeli 2, M. Teshnehlab 3
1Electrical and Computer Eng. Dep, K. N. Toosi University of Technology, Tehran, 16315-1355, Iran
2Faculty of Electrical Engineering, Department of Mechatronics Engineering, Advanced Process Automation & Control Laboratory (APAC), K.N. Toosi University of Technology, Tehran, 16315-1355, Iran
3Faculty of Electrical Engineering, Department of Control Engineering, K. N. Toosi University of Technology, Tehran, 16315-1355, Iran
Correspondence to: F. Keyvanfard , Electrical and Computer Eng. Dep, K. N. Toosi University of Technology, Tehran, 16315-1355, Iran.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Breast cancer Dynamic magnetic resonance imaging (MRI) has emerged as a powerful diagnostic tool for breast cancer detection due to its high sensitivity and has established a role where findings from conventional mammography techniques are equivocal[1]. In the clinical setting, the ANN has been widely applied in breast cancer diagnosis using a subjective impression of different features based on defined criteria. In this study, feature selection and classification methods based on Artificial Neural Network (ANN) and Support Vector Machine (SVM) are applied to classify breast cancer on dynamic Magnetic Resonance Imaging (MRI). The database including benign and malignant lesions is specified to select the features and classify with proposed methods. It was collected from 2004 to 2006. A forward selection method is applied to find the best features for classification. Moreover, several neural networks classifiers like MLP, PNN, GRNN and RBF has been presented on a total of 112 histopathologically verified breast lesions to classify into benign and malignant groups. Also support vector machine have been considered as classifiers. Training and recalling classifiers are obtained with considering four-fold cross validation.
Keywords: Breast MRI, Morphology and Texture Features, Forward Selection, MLP, GRNN, PNN, SVM
Cite this paper: F. Keyvanfard , M. A. Shoorehdeli , M. Teshnehlab , "Feature Selection and Classification of Breast Cancer on Dynamic Magnetic Resonance Imaging Using ANN and SVM", American Journal of Biomedical Engineering, Vol. 1 No. 1, 2011, pp. 20-25. doi: 10.5923/j.ajbe.20110101.04.
Article Outline
1. Introduction
- Breast cancer is the most common cancer and the second leading cause of cancer death in women in Western countries. Imaging has a crucial role in the workup of patients with breast cancer with contributions to early detection through screening, diagnosis and associated image-guided biopsy, treatment planning, and treatment response monitoring [2]. Dynamic contrast-enhanced magnetic resonance (MR) imaging is the most sensitive radiological method for detecting breast cancer [3]. Although, X-Ray mammography has shown considerable success in screening for the early detection of breast cancer, some limitation exist, such as low specificity leading to unnecessary biopsies, presentation of cancer lesions as radio graphically occult in dense breasts, and the inherent limitations of two-dimensional (2D) projection image data. Thus, extensive efforts in recent years have included the use of magnetic resonance imaging (MRI) and sonography as complementary imaging modalities to improve breast-interpretation [2]. Currently, breast MRI has a high sensitivity for breast cancer detection reported as high as 94-100%, but a lower specificity, reported as 37-97%[4].In the clinical reports, the Artificial Neural Network (ANN) has been widely applied in breast cancer diagnosis using different features [5-7]. By presenting examples of known class specification, the neural network is able to learn the distribution patterns of “typical” features of each class (supervised learning). Afterwards, the network can be used to separate non-determined data into the different classes and to calculate the probability for belonging to each of the classes [8]. In this study, a forward selection method is applied to find the best features to the classification of breast cancer. Also, some classifiers are used to classify breast cancer into two groups; benign and malignant lesions.This paper is organized as follows. The database and its quantitative parameters are described in II. Feature selection method and applied classifiers is explained in section III. In section IV, simulation results are summarized on tables to show the performance and the comparison of applied methods. Discussion on the topic is in section V. Finally, the paper is concluded in section VI.
2. Material
2.1. Database
- The database analysed in this study included a total of 112 histology-proven lesions, 86 malignant (41 mass, 45 non-mass) and 26 benign (17 mass, 9 non-mass). These cases were collected from 2004 to 2006. Patients with suspicious findings or with biopsy-proven cancer were invited to participate in a breast MRI research study. Only cases with a clearly visible lesion in a clean background were selected. Cases with multiple poorly differentiable lesions, locally advanced diseases receiving neoadjuvant chemotherapy, or with severe motion artifact, were excluded. The age of patients with malignant cancer ranged from 31 to 80 years (53±11, median 50), and the patients with benign diseases were younger, range 30-74 years old (49±11, median 47). Mass-type lesions were determined based on the clear boundaries and strong enhancements. Lesions presenting as non-mass-like enhancement (or lesions without mass effect) were identified as those presenting diffuse enhancement with ill-defined tumor margins. An experienced radiologist evaluated each lesion and determined whether it should be designated a mass or a non-mass. Table 1 summarizes the histological types of these four groups of lesions. This database was approved in the University of California, Irvine by the institutional review board and in compliance with Health Insurance Portability and Accountability Act (HIPAA) regulations. Written informed consent was obtained from all subjects [9].
2.2. Quantitative Analysis of Lesion Shape Features and Enhancement Texture
2.2.1. Shape Features
- Eight features were used to describe the shape of a lesion: volume, surface area, compactness, NRL (normalized radial length) mean, sphericity, NRL entropy, NRL ratio and roughness. Compactness is defined as the ratio of the square of the surface area to the volume of the lesion—with a sphere having the lowest compactness index and an irregular undulating shape, such as a spiculated lesion, having a higher compactness index. The features based on the normalized radial length (NRL) describe contours and the finer shape of the lesion. NRL is defined as the Euclidean distance from the object’s centre (centre of mass) to each of its contour pixels and normalized relative to the maximum radial length of the lesion[9, 10].
2.2.2. Texture Features
- Radio graphically, texture is defined as a repeating pattern of local variations in image intensity, and is characterized by the spatial distribution of intensity levels in a particular area. Haralick et al. defined 10 grey-level co-occurrence matrix (GLCM) enhancement features (energy, maximum probability, contrast, homogeneity, entropy, correlation, sum average, sum variance, difference average and difference variance) to describe texture[11] .These features were used to characterize lesions with and without mass effect.
2.2.3. Kinetic Features
- The contrast enhancement kinetics was measured from the whole tumor ROI, as well as from the hot spot within the ROI. The hot spot was automatically searched within the whole tumor ROI as the area of 3×3 pixels showing the strongest enhancement on the subtraction image at 1-min after contrast injection. Mean signal intensity was calculated by averaging over the nine pixels of the hot spot ROI, or all pixels within the whole tumor ROI. The percentage enhancement was calculated as the increased signal intensity at each post-contrast frame normalized by the pre-contrast signal intensity (the averaged signal from all four time frames before the injection of contrast agent). The percentage enhancement time course was fitted to a two-compartmental pharmacokinetic Tofts model to characterize the uptake of contrast material in the lesion. The transfer constant (Ktrans) represents contrast uptake in percentage/minute, and the rate constant (kep) captures the washout rate in units of 1/min [9].Table 2 is shown quantitative parameters extracted from ROI.
|
3. Methodology
- For automatic classification of breast cancer on Dynamic MR Imaging, several classifier methods have been combined to separate malignant and benign tumor. This part is described in two sections: feature selection and classifiers.
3.1. Feature Selection
- To select the best features, which produce the least error rate, a forward search strategy is applied. This strategy is between all possible conditions. To control for over-fitting, the potential feature set was limited to no more than five in the all quantitative parameters [9].
3.2. Classifiers
- Several classifiers such as MLP, PNN, GRNN and Support Vector Machine (SVM) have been applied to separate the malignant and benign tumors. These classifiers are introduced in this section.
3.2.1. MLP
- A three-layer feed forward ANN called Multi Layer Perceptron (MLP) has been applied in this paper. In the input layer, the number of nodes is corresponded to the number of input variables. The output node represents 1 and 0 for malignant lesion and benign ones, respectively. Linear and hyperbolic tangent sigmoid transfer functions have been considered as activation functions of input and hidden layers, respectively.Gradient Descent (GD) back propagation and Levenberg-Marquardt (LM) back propagation are applied to train MLP neural network. In GD, the value of weight update is calculated as follows:
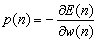 | (2) |
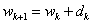 | (3) |
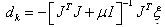 | (4) |
3.2.2. GRNN
- General Regression Neural Network (GRNN) is one of the neural networks applied in this paper. A GRNN can be thought of as a normalized Radial Basis Function (RBF) network in which there is a hidden unit centered at every training case. These RBF units are known as kernels and are usually probability density functions (e.g., Gaussian). The hidden-to-output weights are the target values, so the output is a weighted average of the target values of training cases close to the given input case. The following formula can be used to find the estimated value of random variable Y in any point of space X[13]:
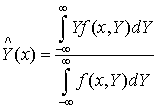 | (5) |
 | (6) |
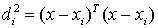 | (7) |
3.2.3. PNN
- Probabilistic Neural Network (PNN) is also applied as a classifier in this paper. The probability density functions of PNN like GRNN are evaluated using the Parzen’s nonparametric estimator. PNN utilizes one probability density function for each category, as shown by (8).
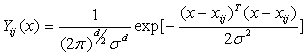 | (8) |
3.2.4. RBF
- A RBF network is a one-hidden-layered feed forward neural network. The first layer consists of input nodes connected to a hidden layer via a set of unity connections, and the outputs of the input layer are determined by calculating the distance between the network inputs and hidden layer centers. A RBF network with no outputs and nh hidden nodes can be expressed as:
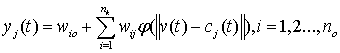 | (9) |
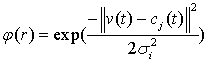 | (10) |
3.2.5. SVM
- The Support Vector Machine (SVM) introduced by V. Vapnik in 1995 is a method to estimate the function classifying the data into two classes[17, 18]. The basic idea of SVM is to construct a hyper plane as the decision surface in such a way that the margin of separation between positive and negative examples is maximized. The SVM term come from the fact that the points in the training set which are closest to the decision surface are called support vectors [19].When used in classification; SVM maps the input space to higher dimensional feature space and constructs a hyper plane, which separates class members from non-members [20].Training vectors xi is mapped into a higher dimensional space by a function Ø. Then SVM finds a linear separating hyper plane with the maximal margin in this higher dimensional space. Furthermore, k (xi, xj) =Ø (xi) Ø (xj) is called the kernel functions. There are number of kernels that can be used in SVM models. These include linear polynomial, RBF and sigmoid[21]:
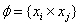 Linear
Linear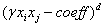 Polynomial
Polynomial 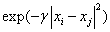 RBF
RBF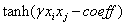 Sigmoidwhere γ> 0 is a parameter that also is defined by the user [21].
Sigmoidwhere γ> 0 is a parameter that also is defined by the user [21].3. Results
- First, every feature of all lesions is normalized to have zero mean and unit variance according to (11). Forward selection algorithm is used for best feature selection. Then selected features are applied to classifiers. To obtain optimal structure of neural networks, the number of hidden units is varied from 2 to the number of input nodes. Therefore, the structure with the lowest validation error is considered as an optimal structure.
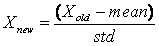 | (11) |
|
|
|
4. Discussion
- Breast cancer is the most common malignancy in the world affecting the female population. The important (primary) aim of this study was to compare several classifiers and to select the best quantitative features to differentiate between malignant and benign breast lesions in patients. Using our data set of 112 lesions, including mass and non-mass-like lesions, forward selection method has been applied for the best feature selection among twenty extracted features. To classify data set into malignant and benign group, several classifiers have been applied in three group data: Mass, Non-mass and all malignant and benign lesions. As shown in above tables, in all type of data, PNN obtained maximum accuracy among all classifiers. According to Figure 1, Ktrans, sum average and entropy are the most important feature in mass group. As you see in Figure 2, sum variance is the most important feature in non-mass group. Also, circularity is significant in classification of non-mass data. According to Figure 3, sum average, homogeneity and correlation, have the greatest number of selection in all data for classification.
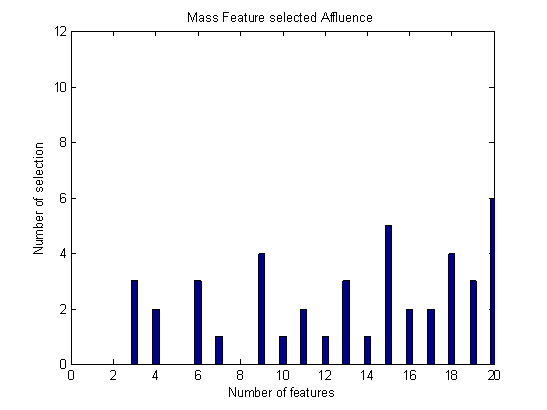 | Figure 1. Number of each feature selection in mass group. |
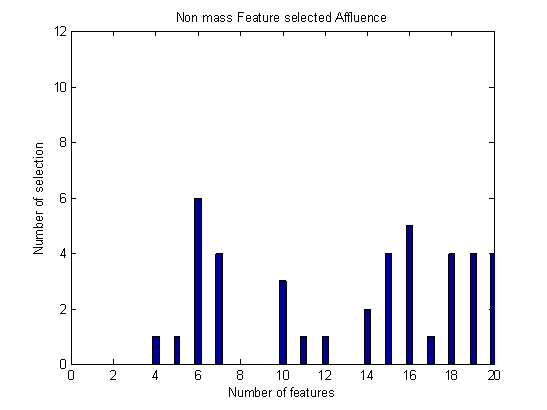 | Figure 2. Number of each feature selection in non-mass group. |
5. Conclusions
- In this study, breast cancer on dynamic MRI is classified by using forward selection and some classification methods. Appropriate quantitative parameters, can be chosen as the best features for classification based on forward selection method. MLPNN, PNN, GRNN, RBF and SVM are applied to classify breast cancer into two groups; benign and malignant lesions. Results confirm that PNN is the best classifier among other methods for classification of these data.
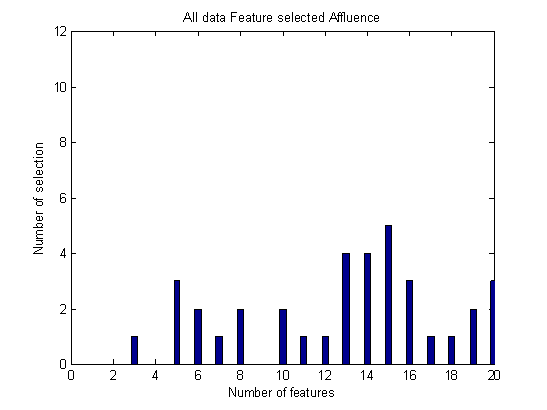 | Figure 3. Number of each feature selection in all data |
ACKNOWLEDGEMENTS
- The authors would like to thank Dr. Min-Ying Su and Ke Nie (University of California) for providing applied database in this work.
References
| [1] | G. P. Liney, M. Sreenivas, P. Gibbs, R. G. Alvarez, and L. W. Turnbull, “Breast lesion analysis of shape technique: Semiautomated vs. manual morphological description” J. Magnetic Resonance Imaging, vol. 23, pp. 493–498, 2006 |
| [2] | W. Chen, L. Giger, U. Brick, “A fuzzy C-Means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR imaging”, Academic Radiology, vol. 13, No.1, pp.63-72, 2006 |
| [3] | B. K. Szabo, M. K. Wiberg, B. Bone, P. Aspelin, “Application of artificial neural networks to the analysis of dynamic MR imaging features of the breast”, Eur Radiol, vol. 14, pp. 1217-1225, 2004 |
| [4] | L. Liberman, EA. Morris, MJ. Lee, JB. Kaplan, LR. LaTrenta, JH. Menell, AF. Abramson, SM. Dashnaw, DJ. Ballon, DD. Dershaw, “Breast lesions detected on MR Imaging: Features and positive predictive value”, J. American Roentgen Ray Society, vol. 179, pp. 171-178, 2002 |
| [5] | L. Arbacha, A. H. Stolpenb, K. S. Berbaum, L. L. Fajardo, J. M. Reinhardt, “Breast MRI lesion classification: Improved performance of human readers with a backpropagation neural network computer- aided diagnosis (CAD) system”, J. Magnetic Resonance Imaging, vol. 25, pp. 89-95, 2007 |
| [6] | M. Nirooei, P. Abdolmaleki, A. Tavakoli, M. Gity, “Feature selection and classification of breast cancer on dynamic magnetic resonance imaging using genetic algorithm and artificial neural networks”, J. Electrical systems, vol. 5, 2009 |
| [7] | P. Abdolmaleki, L D. Buadu, H. Naderimanesh, “Feature extraction and classification of breast cancer on dynamic magnetic resonance imaging using artificial neural network”, Elsevier, Cancer letters, vol.171, pp.183-191, 2001 |
| [8] | R. Lucht, S. Delorme, G. Brix, “Neural network-based segmentation of Dynamic MR mammographic images”, Magnetic Resonance Imaging, vol. 20, pp. 147-154, 2002 |
| [9] | D. Newel, K. Nie, J-H. Chen, C-C. Hsu, H-J. Yu, O. Nalcioglu, M-Y. Su, “ Selection of diagnostic features on breast MRI to differentiate between malignant and benign lesions using computer-aided diagnosis: difference in lesions presenting as mass and non-mass-like enhancement, vol.20, No. 4, pp. 771-781, 2009 |
| [10] | Gibbs P, Turnbull LW “Textural analysis of contrast-enhanced MR images of the breast”, Magn Reson Med 50:92–98. doi:10.1002/mrm.10496, 2003 |
| [11] | Haralick RM, Shanmugam K, Dinstein I (1973) Texture features for image classification. IEEE Trans SMC 3:610– 621. doi:10.1109/TSMC.1973.4309314 |
| [12] | S. Haykin, Neural Networks: A Comprehensive Foundation, 2nd edition, 1999 |
| [13] | E. Al-Daoud, “A comparison between three neural network models for classification problems”, Journal of artificial intelligence, vol. 2, pp. 56-64, 2009 |
| [14] | K. Z. Mao, K. C. Tan, and W. Ser, “Probabilistic Neural-Network Structure Determination for Pattern Classification”, IEEE Trans on NEURAL NETWORKS, vol. 11, No. 4, pp. 1009-1016, 2000 |
| [15] | T. Niwa, “Using general regression and probabilistic neural networks to predict human intestinal absorption with topological descriptors derived from two-dimensional chemical structures”, J. Chem. Inf. Comput. Sci, Vol. 43, pp. 113-119, 2003 |
| [16] | M. Y. Mashor, S. Esugasini, N.A. Mat Isa and N.H. Othman, “Classification of Breast Lesions Using Artificial Neural Network”, IFMBE Proceedings 15, pp. 45-49, 2007 |
| [17] | Haykin, S.: Redes Neurais: Princ´ ıpios e Pr´ atica, 2nd edn. Bookman, Porto Alegre, 2001 |
| [18] | Burges, C. J. C.: A Tutorial on Support Vector Machines for Pattern Recognition. Kluwer Academic Publishers, Dordrecht 1998. |
| [19] | Leonardo de Oliveira Martins, Erick Corrˆ ea da Silva1, Arist´ ofanes Corrˆ ea Silva1, Anselmo Cardoso de Paiva, and Marcelo Gattass, “Classification of Breast Masses in Mammogram Images Using Ripley’s K Function and Support Vector Machine”, Machine Learning and Data Mining in Pattern Recognition, Lecture Notes in Computer Science, Volume 4571, pp. 784-794, 2007 |
| [20] | T. S. Subashini, V. Ramalingam, S. Palanivel, “Breast mass classification based on cytological patterns using RBFNN and SVM”, Expert Systems with Applications, Vol. 36, pp. 5284–5290, 2009 |
| [21] | Y. Ireaneus Anna Rejani, Dr. S. Thamarai Selvi, “EARLY DETECTION OF BREAST CANCER USING SVM CLASSIFIER TECHNIQUE”, International Journal on Computer Science and Engineering Vol.1 (3), pp. 127-130, 2009 |
| [22] | Ron Kohavi, “A Study of Cross-Validation and Bootstrap for Accuracy Estimation and Model Selection”, International Joint Conference on Artificial Intelligence (IJCAI), 1995 |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML