Nalgaa Abou-Elfattah Tawfik1, Omnia Ahmed El-Dydamoni2, Sara Younes Abozaid3, Eman E. Ebrahem4, Marwa Mohamed M. A. Abd EL Rahim5
1Department of Internal Medicine, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
2Department of Medical Microbiology and Immunology, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
3Department of Clinical Pathology, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
4Department of Medical Biochemistry, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
5Department of Rheumatology and Rehabilitation, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt
Correspondence to: Nalgaa Abou-Elfattah Tawfik, Department of Internal Medicine, Faculty of Medicine (for Girls), Al-Azhar University, Cairo, Egypt.
| Email: |  |
Copyright © 2019 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Abstract
Background: Systemic lupus erythematosus (SLE) is an autoimmune disease that often affects women of childbearing age. SLE involves in almost all organs and systems including skin, vascular, joints, and kidneys. The kidney is one of the frequently and seriously affected organs, so lupus nephritis (LN) is a major complication of systemic lupus erythematosus. miRNAs play an important role in kidney physiology and pathology. Objective: To evaluate levels miRNA-146a and miRNA-155 in patients with lupus nephritis and their importance as diagnostic and prognostic biomarkers and their association with disease severity. Patients and Methods: Fifty-six participants were divided into two groups. Group I: twenty-eight systemic lupus nephritis patients taken from rheumatology outpatient clinic and inpatient Internal Medicine Department, Al-Zahraa University Hospital, in the period between July 2018 to April 2019. Group II: twenty-eight healthy volunteers served as a control group with matched age & sex after taking written consent from all patients and controls regarding the ethical committee of the university. miRNA-146a, miRNA-155, C4, C3, ESR, CRP, ANA, DNA, 24H proteins, SLE Disease Activity Index score (SLEDAI) were assayed. Results: The present study revealed that miRNA-146a levels showed a high statistically significant difference (P < 0.001) between cases and controls, also miRNA-155 has statistically significant difference (P= 0.003) between cases and controls, also there were a highly significant positive correlation between miRNA-146a and miRNA-155 (P= 0.000), platelet count (P=0.005); significant positive correlations between miRNA-146a and C4 (P= 0.035), C3 (P= 0.042), 24H urine protein (P= 0.026), creatinine clearance (P= 0.024) and SLEDAI (P= 0.047), also there were significant positive correlations between miRNA-155 and C4 (P= 0.037), C3 (P= 0.039), 24H urine protein (P= 0.046) and SLEDAI (P= 0.039) in lupus nephritis patients. Conclusion: miRNA-146a and miRNA-155 were observed to have a significant correlation with other clinical biochemistry markers, as well as to be prognostic markers for lupus nephritis patients. Also, miRNA-146a and miRNA-155 may serve as a useful specific biomarker for the detection of lupus nephritis among lupus patients in the future, regardless of serum albumin levels and spot urine protein/creatinine ratio.
Keywords:
Lupus nephritis, SLE Disease Activity Index score (SLEDAI), miRNA-146a, miRNA-155
Cite this paper: Nalgaa Abou-Elfattah Tawfik, Omnia Ahmed El-Dydamoni, Sara Younes Abozaid, Eman E. Ebrahem, Marwa Mohamed M. A. Abd EL Rahim, Serum miRNA-146a and miRNA-155 as Novel Biomarkers in Lupus Nephritis Activity with Systemic Lupus Erythematosus, American Journal of Biochemistry, Vol. 9 No. 2, 2019, pp. 21-34. doi: 10.5923/j.ajb.20190902.02.
1. Introduction
Systemic lupus erythematosus (SLE) is a complex autoimmune disease with multi-organ dysfunction [1]. SLE involves in almost all organs and systems, like kidney, skin, joint, vascular [2]. Serum markers of SLE include anti-double-stranded DNA antibodies (anti-dsDNA), anti-C1q antibodies, antinuclear antibodies (ANA), frequently anti-Sm and circulating levels of complement factors such as C3 and C4. These markers are useful for diagnosis but limited for monitoring renal activity [3,4]. 74% of patients with lupus will develop LN [5]. Lupus nephritis (LN) is a serious and most common complication between individuals suffering from SLE and is associated with high morbidity and mortality rates in patients with SLE [5]. Lupus nephritis is a complex process that causing deposition of autoantibodies in the glomerulus, activation of complements and macrophages, cell proliferation, and production of proinflammatory cytokines and chemokines, which are causing tubular damage, tubulointerstitial inflammation, and fibrosis [6]. Current laboratory markers for LN such as proteinuria, urine protein-to-creatinine ratio, creatinine clearance [7]. MicroRNAs (miRNAs) are small noncoding, single-stranded RNA molecules that regulate gene expression at the post-transcriptional level by degrading or blocking translation of messenger RNA (mRNA) [8]. Several of miRNAs could be found in the progression of many diseases [9]. Expression patterns in a few miRNAs in peripheral blood mononuclear cells (PBMCs) from SLE patients compared to healthy controls [10]. The varied expression of miRNAs in kidney makes miRNAs a valuable new tool for understanding, diagnosing, and discovering therapeutic options for SLE and lupus nephritis [7]. miRNA-155 has long been established as a regulator of B-cell functions [11]. miRNA-155 might take part in regulating the production of autoantibodies in SLE. Ablation of miRNA-155 in MRL-lpr lupus-prone mice reduced autoantibody production with the alleviation of kidney inflammation [12]. Another promising miRNA is miRNA-146a, which is recognized as a major negative regulator of immune response, and its deficiency led to multiple immune disorders [13]. The expression of miRNA-146a was shown to correlate with the SLE disease activity and IFN signaling by targeting IRF5 and STAT1 which were both described as important genetic factors in the development of SLE [14]. Elevated levels of miRNA-146a in the kidneys are reported in individuals developing lupus nephritis [15].The aim of this study was to evaluate levels of miRNA-146a and miRNA-155 in patients with lupus nephritis and its importance as diagnostic and prognostic biomarkers and their association with disease severity. Also, it may act as a promising drug in future renal therapy.
2. Patients and Methods
This study was an observational analytical case-control study, was conducted on fifty-six participants; all were females, which were divided into two groups. Group I: twenty-eight systemic lupus nephritis patients. All patients under treatment like corticosteroid, azathioprine, cyclophosphamide or mycophenolate mofetil. SLE was diagnosed according to the American College of Rheumatology (ACR). 2010 ACR/EULAR4 with biopsy-proven nephritis. They were taken from rheumatology outpatient clinic and inpatient Internal Medicine Department, Al-Zahraa University Hospital, in the period between July 2018 to April 2019. Group II: twenty-eight healthy volunteers served as a control group with matched age & sex, after taking written consent from all patients and controls regarding the ethical committee of the university.
2.1. Exclusion Criteria
Patients with other autoimmune diseases like rheumatoid arthritis, scleroderma, overlapping syndrome, pregnant or nursing patients and diabetes mellitus were excluded.All patients were subjected to the following: Full assessment of history and full clinical examination, complete blood count with differential count, fasting blood glucose, kidney function tests (urea & creatinine), liver function tests, lipid profile (cholesterol & triglycerides), coagulation profile, ESR, CRP, 24H protein, creatinine clearance, urine analysis,C4, C3, ANA, (anti-dsDNA), SLE Disease Activity Index score (SLEDAI), miRNA-146a and miRNA-155.
2.2. Quantification of Mature miRNA-146a and miRNA-155 Expression Levels by Real-time PCR (Q-PCR)
To quantify the relative levels of expression of miRNA-146a and miRNA-155 of patients and controls, Quantitative Real-Time Polymerase chain reaction (Q-PCR) was utilized using AB Applied Biosystems Step One Plus, Thermo, Inc., Foster City, CA (USA) according to manufacturer of instructions.
2.2.1. RNA Extraction and Purification
Total RNA was extracted from serum samples using the miRNeasy Mini Kit (Qiagen, Hilden, Germany, Cat. No. 217004) following the manufacture's recommendations. In brief 200 μl of serum sample was mixed with 1 ml QIAzollysis reagent (Lot No. 557010548) and incubated for 5 minutes at room temperature. The 200 μl of chloroform (Sigma) was added, and the tube was vortexed for 15 seconds. After 3 minutes incubation at room temperature, the tube was centrifuged for 15 minutes at 14000 rpm at 4°C. 600 μl of upper aqueous phase was transferred to a new collection tube. 900 μl of 99.9% ethanol was added and mixed thoroughly by pipetting. 700 μl of sample was transferred into RNeasy Min column in a 2 ml collection tube and centrifuged for 15 seconds at 10000 rpm. Discard the flow-through. RNeasy Min column was washed one time by buffer RWT and two times by buffer RPE, and 30-50 μl of RNase-free water were pipetted directly onto the Rneasy membrane and centrifuged for 1 min at 10000 rpm to elute RNA. The RNA purity in the extract was assessed by the Nanodrop Spectrophotometer, ThermoFisher.
2.2.2. Reverse Transcription
Purified RNA was then used for one-step reverse transcription using miScript II RT kit (Qiagen, Hilden, Germany, Cat. No. 218161, Lot no. 163012117) following the manufacturer's instruction. Briefly, 4 μl of 5x miScript HiSpec buffer, 2 μl of 10x miScript nucleic Mix was mixed with 2 μl of miScript reverse transcriptase mix. 12 μl template of RNA was added to each 0.2 PCR tube with total reaction volume 20 μl / reaction. After gentle mix, the tubes are placed in thermal cycler (Bio-Rad, T100, Singapore) and the run condition was adjusted as follows: incubate for 60 minutes at 37°C followed by incubation for 5 min at 95°C to inactivate miScript reverse transcriptase mix.
2.2.3. cDNA Dilution Prior to PCR
Ten μl of concentrated cDNA was added to 50 μl of nuclease-free water to start PCR reaction immediately.
2.2.4. PCR Quantification of Mature miRNA-146a and miRNA-155
PCR quantification was performed with real-time PCR using the SYBR Green PCR Master Mix (Qiagen, Hilden, Germany, Cat. No. 218073) according to manufacturer’s protocol. Briefly, three tubes of master mix were prepared contained 12.5 μl of SYBR Green PCR Master Mix, 2.5 μl of universal primer and 2.5 μl nuclease-free water. 2.5 μl forward primer miRNA146a-5 p (Cat. No. MS00003535) was added to first tube followed by 2.5 μl forward primer miRNA155-5 p (Cat. No. MS00031486) was added to second tube, and 2.5 μl housekeeping gene SNORD 68 primer (Cat. No. MS00033712) was added to third tube. Five μl of diluted cDNA template were added to each tube, with total reaction volume 25 μl/ reaction. The reaction tubes were subjected to the following conditions: an initial activation step at 95°C for 15 min followed by 40 cycles with denaturation 94°C for 15 seconds, annealing at 55°C for 30 seconds and extension at 70°C for 30 seconds. Fluorescence readings were taken after each cycle. Melting curve analysis of PCR products at 65-95°C to verify their specificity and identity.All primers used for the amplification and housekeeping gene primer were supplied by (Qiagen, Hilden, Germany) are listed in the table (1) and figure (1).Table 1. Primer sequence of miRNA146a and miRNA155
 |
| |
|
 | Figure 1. Amplification blots of clinical samples that expressed miRNA-146a and miRNA-155 |
2.2.5. PCR Results Analysis
The ∆ Ct was calculated by subtracting the Ct values of miRNA SNORD 68 from the Ct values of the target miRNA 146a-5p and miRNA 155-5p. The resulting normalized ∆ Ct values were used in calculating relative expression values of individual miRNAs by using 2-∆∆ Ct method these values are directly related to the miRNA expression levels.
2.3. Statistical Analysis
Statistical analysis was done using the Statistical Package for the Social Sciences (SPSS software version 25, Chicago, Illinois). Quantitative data were expressed as mean ± standard deviation (SD). Mean, (±SD) and range for parametric numerical data, while Median (IQR) was used for non-parametric numerical data. Qualitative data were expressed as frequency and percentage. Student t-test was used to assess the statistical significance of the difference between two study group means values of quantitative data. Chi-Square test was used to compare the relationship between two qualitative variables. Mann Whitney Test (U test) was used to assess the statistical significance of the difference of a non-parametric variable between two study groups. The receiver operating characteristic (ROC) curves were used to determine the cutoff point, which shows the highest sensitivity and specificity of miRNA-146a and miRNA-155 in differentiating between different conditions.
3. Results
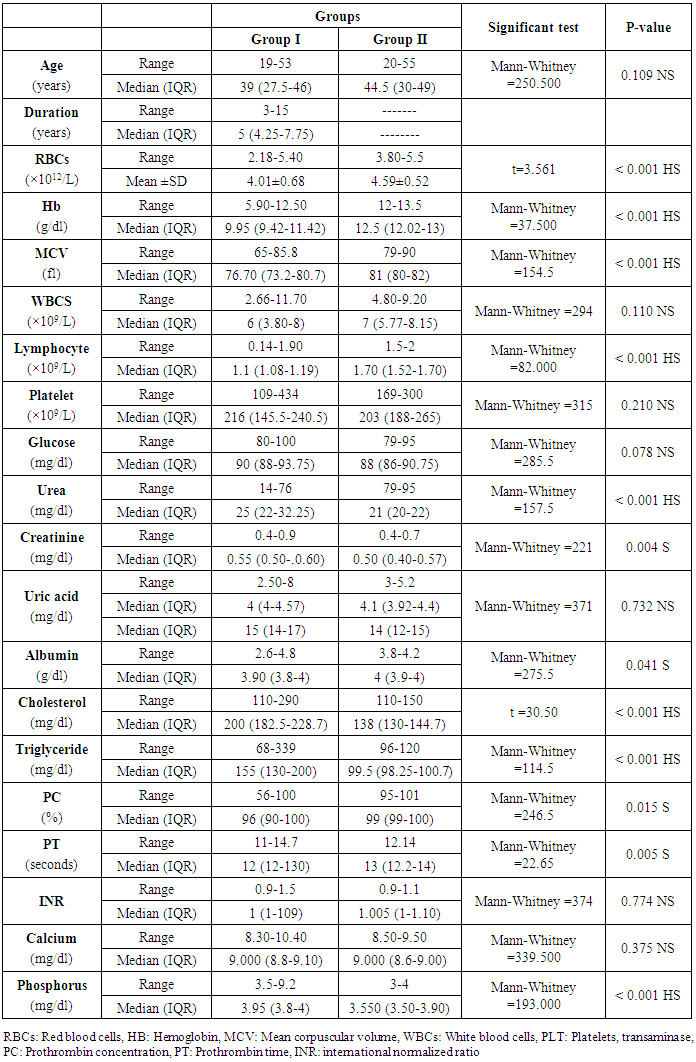
This study included two groups: patients' group (twenty-eight SLE female patients, 100%), compared to the control group (twenty-eight healthy female subjects, 100%). The demographic data and laboratory findings of SLE patients and controls included in the study; their age ranged from 19-53 years, with median (IQR) 39 years and control group their age ranged from 20-55 years, with median (IQR) 44.5. There were highly statistically significant differences between cases and controls as regards RBCs, Hb, MCV, lymphocyte, urea, cholesterol, triglycerides, phosphorus, their levels were detected in the patients with a mean ± SD of (4.01±0.68) for RBCs, median (IQR) of 9.95 for Hb, median (IQR) of 76.70 for MCV, median (IQR) of 1.1 for lymphocyte, median (IQR) of 25 for urea, median (IQR 200 for cholesterol, median (IQR) of 155 for triglycerides, median (IQR) of 3.95 for phosphorus, while their levels were detected in the controls with a mean ± SD of (4.59±0.52) for RBCs, median (IQR) of 12.5 for Hb, median (IQR) of 81 for MCV, median (IQR) of 1.70 for lymphocyte, median (IQR) of 21 for urea, median (IQR) 138 for cholesterol, median (IQR) of 99.5 for triglycerides, median (IQR) of 3.550 for phosphorus. Also, there were statistically significant differences between cases and controls as regards creatinine, albumin, PC, and PT, their levels were detected in the patients with a median (IQR) of 0.55 for creatinine, median (IQR) of 3.90 for albumin, median (IQR) of 96 for PC and median (IQR) of 12 for PT; while their levels were detected in the controls with a median (IQR) of 0.50 for creatinine, median (IQR) of 4 for albumin, median (IQR) of 99 for PC and median (IQR) of 13 for PT (Table 2).Table 2. Demographic data and laboratory findings of SLE patients and controls
 |
| |
|
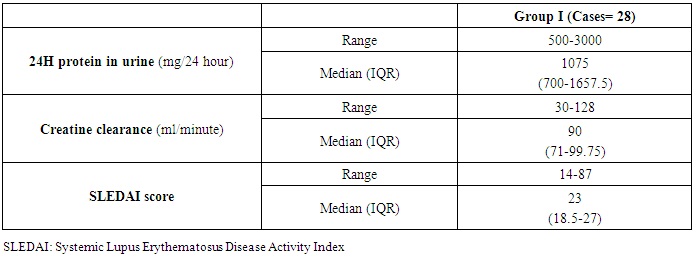
The 24H Protein, creatinine clearance, and SLEDI score were calculated in the patient group with a mean and a median (IQR) of (500-3000) and 1075, (30-128) and 90, (14-87) and 23, respectively (Table 3).Table 3. Renal parameters and SLEDI of lupus nephritis patients
 |
| |
|
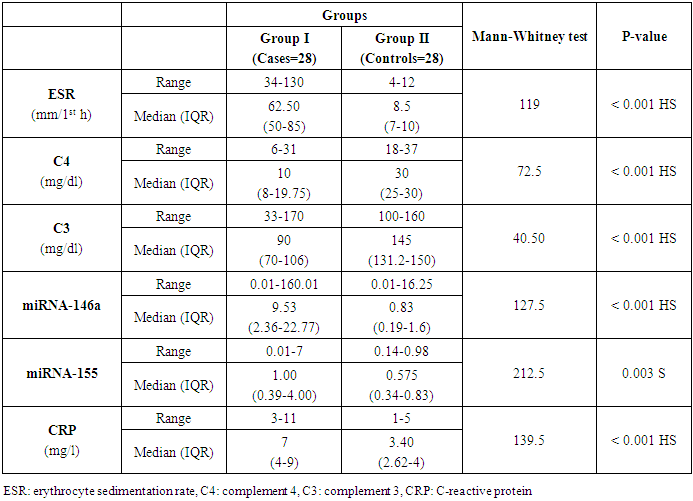
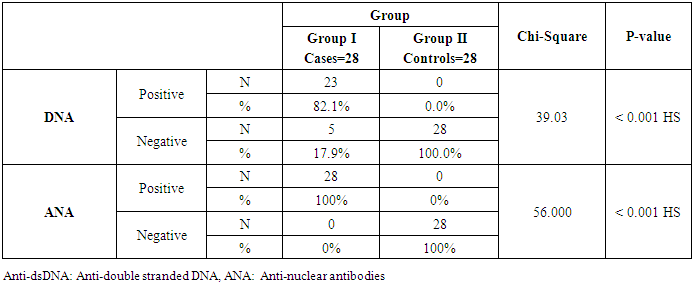
Both groups were compared according to certain laboratory markers. ESR, C4, C3, miRNA-146a and CRP levels showed a high statistically significant difference (P < 0.001) and miRNA-155 showed statistically significant difference (P= 0.003) between cases and controls with an ESR range and a median (IQR) of (34-130) and 62.5 in patients and (4-12) and 8.5 in controls. C4 range and a median (IQR) of (6-31) and 10 in patients and (18-37) and 30 in controls. C3 range and a median (IQR) of (33-170) and 90 in patients and (100-160) and 145 in controls. miRNA-146a range and a median (IQR) of (0.01-160.01) and 9.53 in patients and (0.01-16.25) and 0.83 in controls. CRP range and a median (IQR) of (3-11) and 7 in patients and (1-5) and 3.4 in controls. miRNA-155 range and a median (IQR) of (0.01-7) and 1 in patients and (0.14-0.98) and 0.575 in controls (Table 4 and Figs. 2 & 3). Anti-dsDNA was positive in 23 patients with a percentage of 81.1% and negative in 5 patients with a percentage of 17.9% but it was not detected (positive = zero = 0%) in all controls (negative = 28 = 100%); while ANA was positive in all 28 patients with a percentage of 100% and negative in all 28 controls (100%) (Table 5).Table 4. Comparison of serum levels of ESR, CRP, C3, C4, miRNA-146a, and miRNA-155 and between patients and controls
 |
| |
|
Table 5. Immunological parameters between patients and controls
 |
| |
|
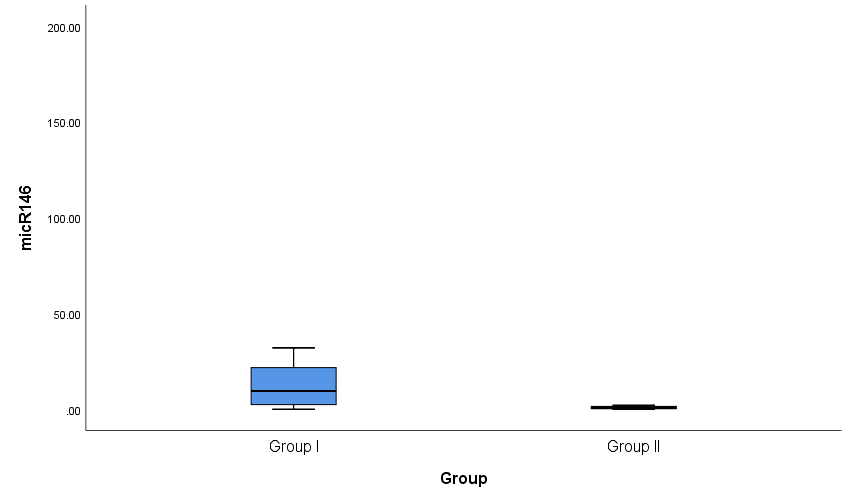 | Figure 2. Box plots for value distribution ofmiRNA-146a in group I and group II |
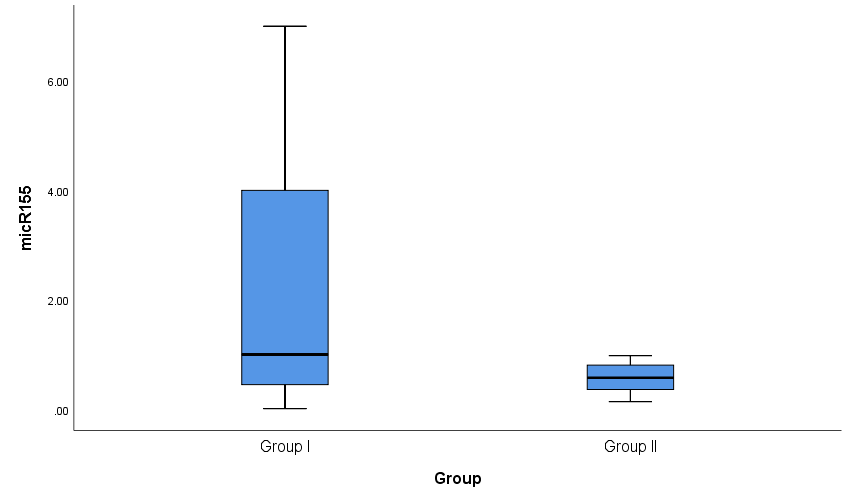 | Figure 3. Box plots for value distribution of miRNA-155 in group I and group II |
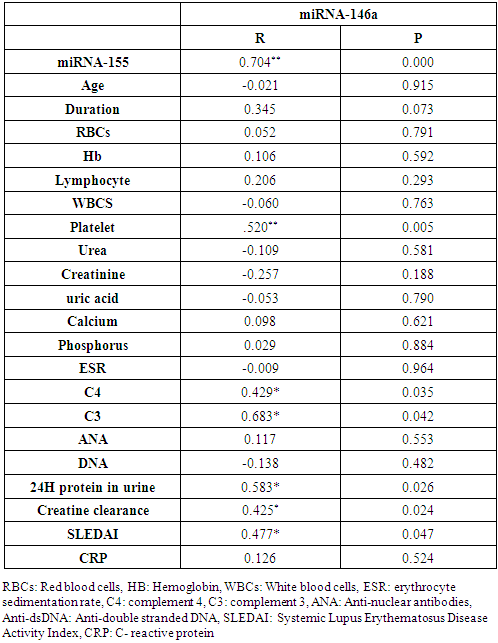
In the correlation of miRNA-146a with miRNA-155 and different studied parameters among lupus nephritis patients, there were a highly significant positive correlation between miRNA-146a and miRNA-155 (P= 0.000), platelet count (P=0.005); significant positive correlations between miRNA-146a and C4 (P= 0.035), C3 (P= 0.042), 24H urine protein (P= 0.026), creatinine clearance (P= 0.024) and SLEDAI (P= 0.047), while there was no significant correlation between miRNA-146a and the age of the patients (P= 0.915), duration of the disease (P= 0.073), RBCs count (P= 0.791), Hb concentration (P= 0.592), lymphocyte count (P= 0.293), WBCs count (P= 0.763), serum urea level (P= 0.581), serum creatinine level (P= 0.188), serum uric acid level (P= 0.790), serum calcium level (P= 0.621), serum phosphorus level (P= 0.884), ESR (P= 0.964), ANA (P= 0.553), anti-dsDNA (P= 0.482), or CRP (P= 0.524) (Table 6 and Figs. 4 & 5).Table 6. Correlation between miRNA-146a and demographic and laboratory data
 |
| |
|
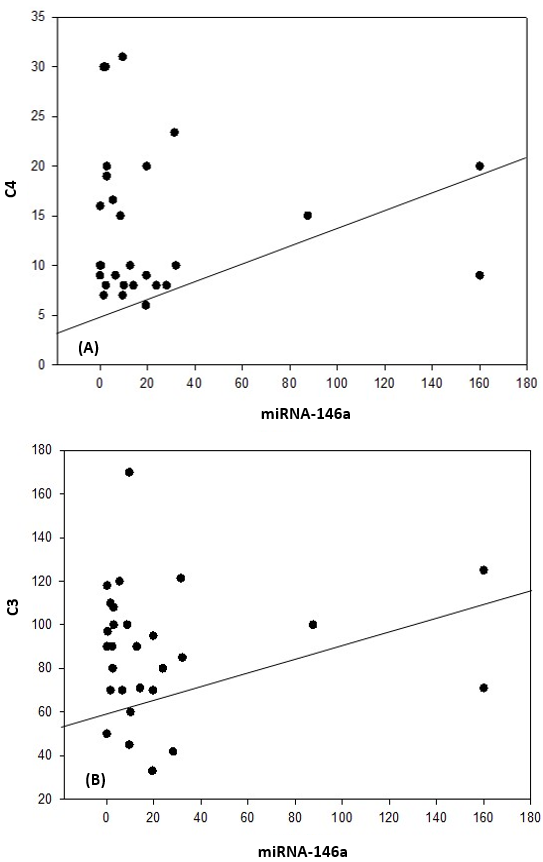 | Figure 4. Correlation coefficient in group 1 (cases). A) between miRNA-146a and C4 (P= 0.035). B) between miRNA-146a and C3 (P= 0.042) |
 | Figure 5. Correlation coefficient in group 1 (cases). A) between miRNA-146a and 24H protein (P= 0.026). B) between miRNA-146a and SLEDAI (P= 0.047) |
In the correlation of miRNA-155 with miRNA-146a and different studied parameters among lupus nephritis patients, there were a highly significant positive correlation between miRNA-155 and miRNA-146a (P= 0.000), significant positive correlations between miRNA-155 and C4 (P= 0.037), C3 (P= 0.039), 24H urine protein (P= 0.046) and SLEDAI (P= 0.039), while there was no significant correlation between miRNA-155 and the age of the patients (P= 0.70), duration of the disease (P= 0.76), RBCs count (P= 0.95), Hb concentration (P= 0.60), lymphocyte count (P= 0.23), WBCs count (P= 0.12), platelet count (P= 0.10), serum urea level (P= 0.91), serum creatinine level (P= 0.90), serum uric acid level (P= 0.16), serum calcium level (P= 0.81), serum phosphorus level (P= 0.67), ESR (P= 0.16), ANA (P= 0.21), anti-dsDNA (P= 0.15) creatinine clearance (P= 0.13) or CRP (P=0.22) (Table 7 and Figs. 6 & 7 & 8).Table 7. Correlation between miRNA-155 and demographic and laboratory data
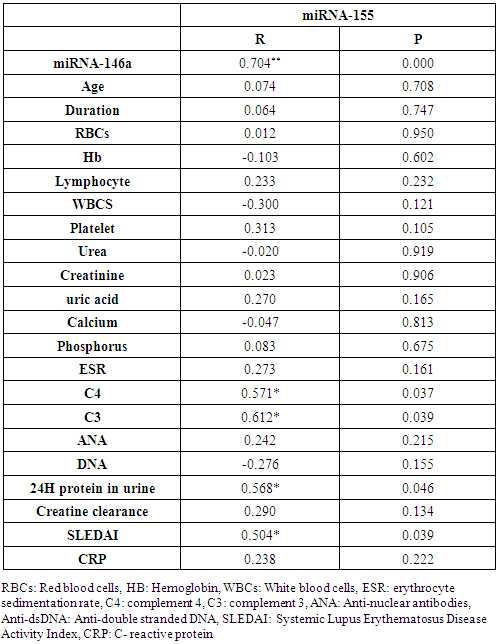 |
| |
|
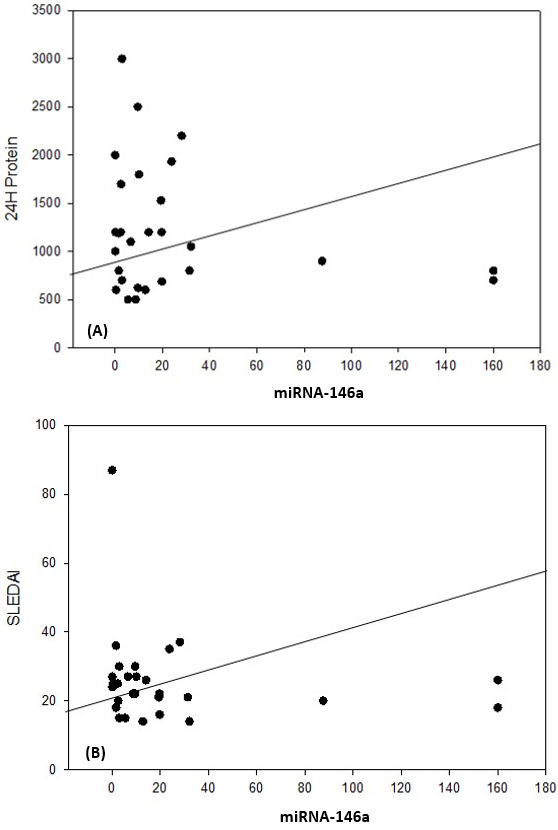
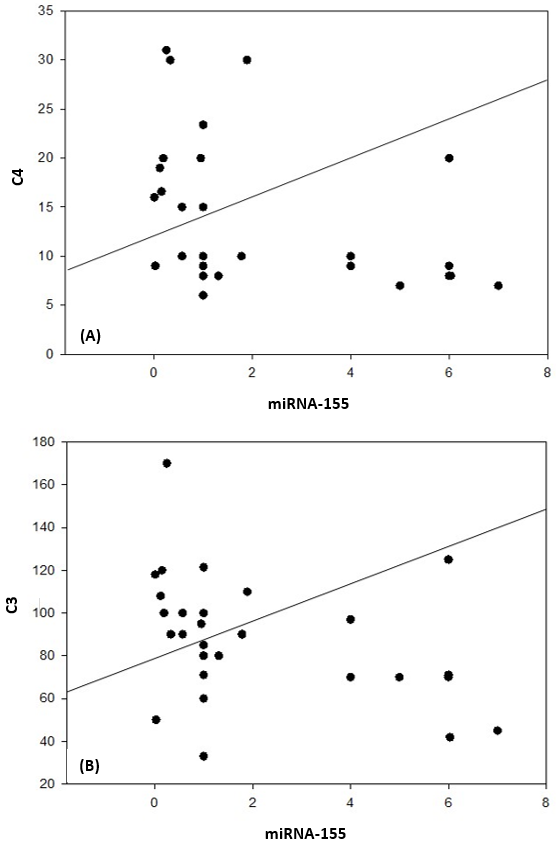 | Figure 6. Correlation coefficient in group 1 (cases). A) between miRNA-155 and C4 (P= 0.037). B) between miRNA-155 and C3 (P= 0.039) |
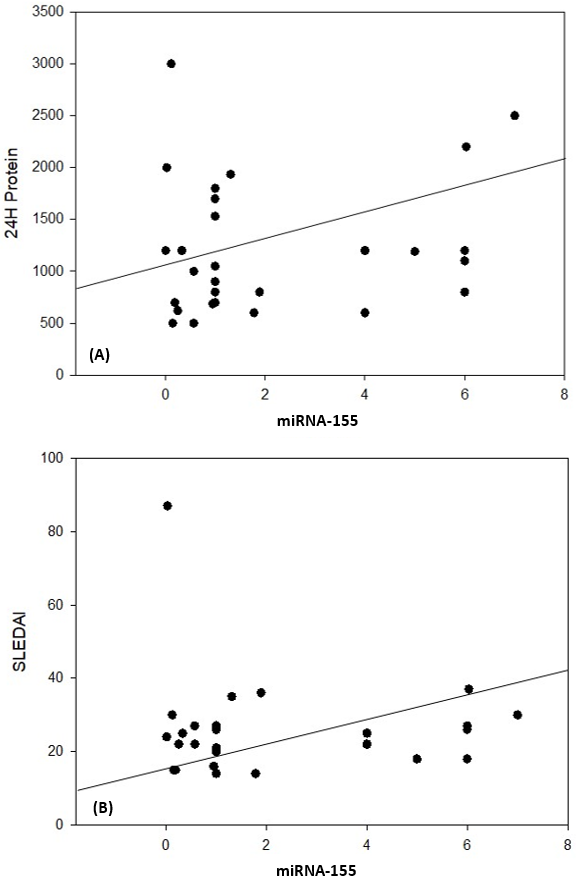 | Figure 7. Correlation coefficient in group 1 (cases). A) between miRNA-155 and 24H protein (P= 0.042). B) between miRNA155 and SLEDAI (P= 0.039) |
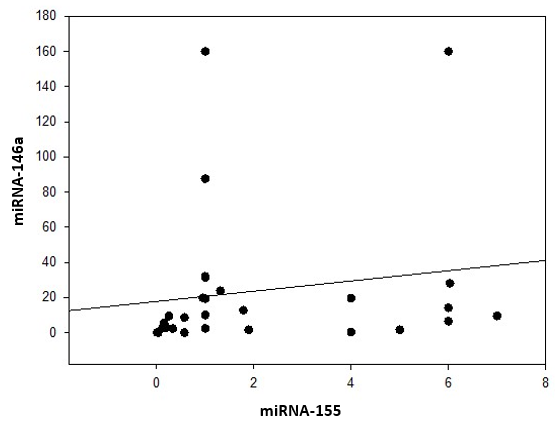 | Figure 8. Correlation coefficient in group 1 (cases) between miRNA-155 and miRNA-146a |
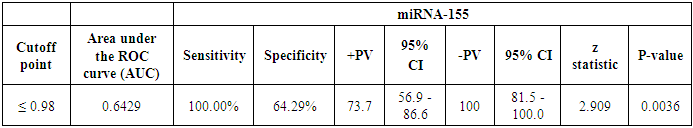
The cutoff point of miRNA-146a for the diagnosis of lupus nephritis was ≤ 2.1 with a sensitivity of 92.86% and specificity of 78.57%, PPV of 81.2% and NPV of 91.7%, 95% CI of (63.6-92.8) (Table 8 and Fig. 9). While the cutoff point of miRNA-155 for the diagnosis of lupus nephritis was ≤ 0.98 with a sensitivity of 100% and specificity of 64.29%, PPV of 73.7% and NPV of 100%, 95% CI of (56.9-86.6) (Table 9 and Fig. 10).Table 8. The cutoff values and their diagnostic significance for themiRNA-146a in LN patients
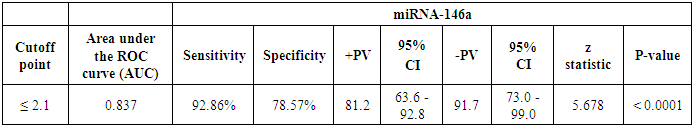 |
| |
|
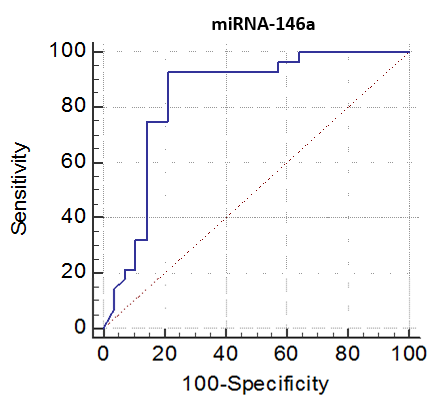 | Figure 9. Receiver operating characteristics (ROC) curve to define the best cutoff values of miRNA-146a |
Table 9. The cutoff values and their diagnostic significance for the miRNA-155 in LN patients
 |
| |
|
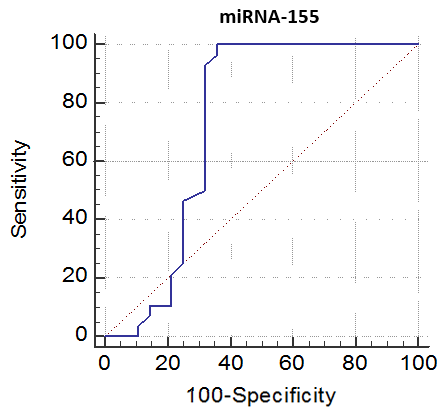 | Figure 10. Receiver operating characteristics (ROC) curve to define the best cutoff values of miRNA-155 |
4. Discussion
The micro-ribonucleic acids (miRNAs) are small non-coding RNAs of 18–25 nucleotides that regulate the expression of multiple protein-encoding genes [16]. miRNAs play a critical role in the regulation of gene expression, development of the immune system and also regulation of both innate and adaptive immunity [17]. Serum and urinary miRNAs were shown to be associated with different disease characteristics of SLE [18]. Expression of miRNA can be used to evaluate the severity of the disease, monitor the patients and changes in the management of the disease [19] elevated levels of miRNA-146a in the kidneys are reported in individuals developing lupus nephritis [20]. In the present study there was increase of miRNA-146a inpatient with lupus nephritis in comparison to healthy control groups this result in agreeing with Labib et al. [21] showed a significantly increased level of miRNA-146a-5p in serum of SLE patients compared to controls. Also, agree with Zununi Vahed et al. [22] who report a possible role for miRNA-146a in the immune system or immunity dysregulation and pathogenesis of LN [22]. Also, disagree with Hashad et al. [23] who report down regulation of miRNA-146a in lupus nephritis. Also, our results disagree with Wang et al. [8] who report down-regulation of miRNA-146a in serum of SLE patients and inversely correlated with proteinuria [8], also disagree with Zhu et al. [24] and Zheng et al. [25]. The difference explained by that miRNA-146a expression is up-regulated in Th1 cells and down-regulated in Th2 cells relative to its expression in naive T cells [26]. Also, in our study, there was increase of miRNA-155 inpatient with lupus nephritis in comparison to healthy control group this result in agree with Zununi Vahed et al. [22]. Explained by due to inflammation which inflammation-mediated glomerular endothelial injury, and fibrosis [27]. Specific miRNAs, such as miRNA-146a, miRNA-155, were initially shown to be upregulated during the macrophage inflammatory response [28]. Our results reported that there was correlation between miRNA-155 and miRNA-146a, C4, C3, 24H protein in urine, SLEDAI, this result in agree with Zununi et al. [22] who reported miRNA-155 was correlated with SLEDAI scores and miRNA-146a. Also, our results revealed there was positive correlation between miRNA-146a and 24H protein, SLEDAI, creatinine clearance, platelet and no correlation with leukocyte, neutrophil, and creatinine levels, this results in disagreeing to Su et al. [29] who report there was positive correlation between miRNA-146a-5p and leukocyte, neutrophil, and creatinine levels, as well as the negative correlation of the lymphocyte percentage with miRNA-146a-5p. However, miRNA-146a-5 is not correlated with the albumin level or the urine protein/creatinine level (both P > 0.05). This finding explains using miRNA-146a-5p in clinical situations as a marker to detect LN even before the albumin level decreases or the urine protein/creatinine level increases. Our result revealed that miRNA-146a with sensitivity and specificity of respectively 92.86% and 78.57%. Also, miRNA-155 a sensitivity and specificity of respectively 100.00% and 64.29%. In comparison to Zununi Vahed et al. [22] reported that miRNA-146a with sensitivity and specificity of respectively 56% and 96%. miRNA-155 presented a sensitivity of 88% and specificity of 67%. Also, results reported by Labib et al. [21] of miRNA-146a sensitivity of 69%, a specificity of 64.1%. The difference between miRNA levels among lupus nephritis patients of our study and in different studies is possibly due to the different ethnicity of patients, exposure to infections and different environmental factors [30]. Also, another explanation small sample size, and different sample sources, lifestyle, dietary habits, and different nephritis therapy.
5. Conclusions
miRNA-146a and miRNA-155 were observed to have a significant correlation with other clinical biochemistry markers, as well as to be a prognostic marker for lupus nephritis patients. Also, miRNA-146a and miRNA-155 may serve as a useful specific biomarker for the detection of lupus nephritis among lupus patients in the future, regardless of serum albumin levels and spot urine protein/creatinine ratio.
References
| [1] | Zeng L, Wu JL, Liu LM, Jiang JQ, Wu HJ, Zhao M and Lu QJ (2018). Serum miRNA-371b-5p and miRNA-5100 act as biomarkers for systemic lupus erythematosus. Clin Immunol, 196: 103-109. |
| [2] | Rothfield N, Sontheimer RD and Bernstein M (2006). Lupus erythematosus: systemic and cutaneous manifestations. Clin Dermatol, 24(5): 348-362. |
| [3] | Manzella DJ, Dettori PN, Hertimian ML and Melero MJ (2013). J Clin Rheumatol, 19(2): 87-89. |
| [4] | Manoharan A and Madaio MP (2010). Biomarkers in Lupus Nephritis. Rheum. Dis. Clin. N. Am., 36: 131–143. |
| [5] | Li Y, Fang X and Li QZ (2013). Biomarker profiling for lupus nephritis. Genomics Proteomics Bioinformatics, 11(3): 158-165. |
| [6] | Gurevitz SL, Snyder JA, Wessel EK, Frey J and Williamson BA (2013). Consult Pharm, 28(2): 110-121. |
| [7] | Pacheco L, Díaz Y and Aroca G (2014). Biomarkers in biological fluids and their potential use as indicators of lupus nephritis in individuals with systemic lupus erythematous. Rev. Colomb. Nefrol., 1(1): 39-47. |
| [8] | Wang G, Tam LS, Li EK, Kwan BC, Chow KM, Luk CC, et al. (2010). Serum and urinary cell-free MiR-146a and MiR-155 in patients with systemic lupus erythematosus. J Rheumatol, 37(12): 2516-2522. |
| [9] | Lian D, Wang ZZ and Liu NS (2016). MicroRNA-1908 is a biomarker for poor prognosis in human osteosarcoma. Eur Rev Med Pharmacol Sci, 20(7): 1258-1262. |
| [10] | Honarpisheh M, Köhler P, von Rauchhaupt E and Lech M (2018). The Involvement of MicroRNAs in modulation of innate and adaptive immunity in systemic lupus erythematosus and lupus nephritis. J Immunol Res, 2018: 4126106. |
| [11] | Thai TH1, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. (2007). Regulation of the germinal center response by microRNA-155. Science, 316(5824): 604-608. |
| [12] | Thai TH, Patterson HC, Pham DH, Kis-Toth K, Kaminski DA and Tsokos GC (2013). Deletion of microRNA-155 reduces autoantibody responses and alleviates lupus-like disease in the Fas(lpr) mouse. Proc Natl Acad Sci U S A, 110(50): 20194-20199. |
| [13] | Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. (2011). miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med, 208(6): 1189-1201. |
| [14] | Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, et al. (2009). MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum, 60(4): 1065-1075. |
| [15] | Ichii O and Horino T (2018). MicroRNAs associated with the development of kidney diseases in humans and animals. J Toxicol Pathol, 31(1): 23-34. |
| [16] | Wu L, Fan J and Belasco JG (2006). MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A, 103(11): 4034-4039. |
| [17] | Dai R and Ahmed SA (2011). MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res, 157(4): 163-179. |
| [18] | Wang G, Tam LS, Li EK, Kwan BC, Chow KM, Luk CC, et al. (2011). Serum and urinary free microRNA level in patients with systemic lupus erythematosus. Lupus, 20(5): 493-500. |
| [19] | Leiss H, Salzberger W, Jacobs B, Gessl I, Kozakowski N, Blüml S, et al. (2017). MicroRNA 155-deficiency leads to decreased autoantibody levels and reduced severity of nephritis and pneumonitis in pristane-induced lupus. PLoS One, 12(7): e0181015. |
| [20] | Lu J, Kwan BC, Lai FM, Tam LS, Li EK, Chow KM, et al. (2012). Glomerular and tubulointerstitial miR-638, miR-198 and miR-146a expression in lupus nephritis. Nephrology (Carlton), 17(4): 346-351. |
| [21] | Labib DA, Koptan D, Ghoniem S, Salah SH, El Shazly R and El Refai RM (2019). Dysregulation of microRNA146a-5p expression in systemic lupus erythematosus females: Diagnostic potential and association with ocular manifestations. The Egyptian Rheumatologist, Available online 19 July 2019. |
| [22] | Zununi Vahed S, Nakhjavani M, Etemadi J, Jamshidi H, Jadidian N, Pourlak T and Abediazar S (2018). Altered levels of immune-regulatory microRNAs in plasma samples of patients with lupus nephritis. Bioimpacts, 8(3): 177-183. |
| [23] | Hashad DI, Abdelmagid MH and Elsherif SH (2012). microRNA146a expression in lupus patients with and without renal complications. J Clin Lab Anal, 26(1): 35-40. |
| [24] | Zhu Y, Xue Z and Di L (2017). Regulation of MiR-146a and TRAF6 in the diagnose of lupus nephritis. Med Sci Monit, 23: 2550-2557. |
| [25] | Zheng CZ, Shu YB, Luo YL, and Luo J (2017). The role of miR-146a in modulating TRAF6-induced inflammation during lupus nephritis. Eur Rev Med Pharmacol Sci, 21(5): 1041-1048. |
| [26] | Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, et al. (2005). MicroRNA profiling of the murine hematopoietic system. Genome Biol, 6(8): R71. |
| [27] | Zununi Vahed S, Poursadegh Zonouzi A, Ghanbarian H, Ghojazadeh M, Samadi N, et al. (2017). Differential expression of circulating miR-21, miR-142-3p and miR-155 in renal transplant recipients with impaired graft function. Int Urol Nephrol, 49(9): 1681-1689. |
| [28] | O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, et al. (2010). MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity, 33(4): 607-619. |
| [29] | Su YJ, Lin IC, Wang L, Lu CH, Huang YL and Kuo HC (2018). Next generation sequencing identifies miRNA-based biomarker panel for lupus nephritis. Oncotarget, 9(46): 27911-27919. |
| [30] | Chung JW, Jeong SH, Lee SM, Pak JH, Lee GH, Jeong JY and Kim JH (2017). Expression of MicroRNA in host cells infected with Helicobacter pylori. Gut Liver, 11(3): 392-400. |













 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML







