-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Biochemistry
p-ISSN: 2163-3010 e-ISSN: 2163-3029
2017; 7(5): 127-133
doi:10.5923/j.ajb.20170705.05

The Association between Interleukin-10 Gene Polymorphism (-1082 G/A) and Bronchial Asthma with the Relation to Spirometric Parameters in Egyptian Children
Radwa S. Shahin1, Rasha M. Gouda2
1Clinical Pathology Department, Faculty of Medicine (for girls), Al-Azhar University, Cairo, Egypt
2Pediatric Department, Faculty of Medicine (for girls), Al-Azhar University, Cairo, Egypt
Correspondence to: Radwa S. Shahin, Clinical Pathology Department, Faculty of Medicine (for girls), Al-Azhar University, Cairo, Egypt.
| Email: |  |
Copyright © 2017 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Background: Asthma is a chronic inflammatory disorder of the airways in which many cells and cellular elements play a role. Bronchial asthma associated with airway hyper-responsiveness, reversible airway obstruction, and allergen-specific IgE production. Growing evidences suggest the association between IL-10 promoter polymorphism and asthma production. Objective: This study was conducted to evaluate the association of IL-10 promoter polymorphism rs1800896 (-1082 G/A) with asthma and its relation to spirometric parameters. Methods: IL-10 polymorphism (rs1800896) was genotyped using real time PCR technique and IgE serum level was measured by ELISA in 30 asthmatic children and 20 controls. Results: There was highly significant increase in IgE level and Eousenophil (%) in asthmatic children compared to control group (p= 0.000 and 0.002). There was significant association between IL-10 polymorphisms (rs1800896) and bronchial asthma.Conclusion: The results of this study suggest that the IL-10 -1082 G/A polymorphism can be associated with susceptibility to pediatric asthma and can be related to reduced respiratory functions between asthmatics.
Keywords: IL-10 polymorphisms (rs1800896), IgE, Bronchial athma, Pulmonary function tests
Cite this paper: Radwa S. Shahin, Rasha M. Gouda, The Association between Interleukin-10 Gene Polymorphism (-1082 G/A) and Bronchial Asthma with the Relation to Spirometric Parameters in Egyptian Children, American Journal of Biochemistry, Vol. 7 No. 5, 2017, pp. 127-133. doi: 10.5923/j.ajb.20170705.05.
Article Outline
1. Introduction
- Bronchial asthma (BA) is heterogeneous disease, usually characterized by chronic airway inflammation [1]. It is defined by history of respiratory symptoms such as wheez, shortness of breath, chest tightness and cough that vary over time and in intensity, together with variable expiratory airflow limitation [2].Asthma is a chronic disease of high prevalence affecting 330 million people world-wide. Evidence show that the prevalence of asthma is increasing, especially in children [3].Pulmonary function tests are a group of tests that measure how well the lungs take in and release air and how well they move gases such as oxygen from the atmosphere into the body’s circulation [4]. Clinical diagnosis of asthma should be supported by demonstration of variability of airway obstruction on pulmonary function testing over a period, even if not demonstrated at a point [5].The development of asthma is determined by the interaction between host genetic susceptibility and environment. Although the etiology of asthma remains unknown, an imbalance between T-helper 1 (Th1)/Th2 paradigm has a dominating genetic component [6].Regulatory T (Treg) cells play a key role in balancing Th1/Th2 level. Treg cells contribute to immune regulating and anti-inflammatory responses due to down-regulation of antigen and macrophage activation through various proteins, including interleukin (IL)-10 [7].IL-10 was first identified in 1989 as cytokine synthesis inhibitory factor which is synthesized by activated CD4 and CD8 T lymphocytes, mast cells and activated monocytes and has a down-regulatory effect on inflammatory responses. The IL-10 gene is located on chromosome 1 (1q31-32) in human genome and is consisted of 5 exons and 4 introns [8].IL-10 is a key factor in the development of immune tolerance with a regulatory role in both inflammatory and anti-inflammatory processes. Polymorphism in IL10 gene affects the level of IL-10 production, so that the carriage of allele G (rs1800896) is associated with higher IL-10 production. Low amount of mononuclear cell secreting IL-10 has increased the risk of later wheezing. IL-10 rs1800896 promoter polymorphism has been linked with the etiology and severity of asthma [9].Elevated serum levels of specific IgE towards common environmental allergens are a key component in the pathogenesis of allergic asthma. IgE antibodies cause chronic airway inflammation through effector cells such as mast cells, basophils etc, activated via high-affinity (FcεRI) or low-affinity (FcεRII) IgE receptors [10].The aim of the present study was to evaluate the associations between bronchial asthma and polymorphism in IL10 rs1800896 (-1082 G/A) in Egyptian children and to study its relation to diminished respiratory functions in asthmatics.
2. Subjects and Methods
- This study was carried out on thirty children suffering from bronchial asthma selected from Al-Zahraa University Hospital, pediatric outpatients clinic and inward pediatric department diagnosed as asthma fulfilling GINA Guidelines 2014. Their ages ranged from 6 and 15 years old (10.3 ± 3.1), there were 12 males and 18 females. Twenty samples were collected from age and gender matched apparently healthy children, to be used as control, their ages ranged from 7 and 15 years old (11.2 ± 2.8), there were 11 males and 9 female. The study was approved by the Ethics Committee of Al-Azhar University and a written informed consent was obtained from children's guardians.All cases and control subjects were subjected to the following procedure:1- Full history taking including symptoms and sings of asthma to fulfill criteria of GINA guidelines including impact of asthma on patient activity and school attendance, also demographic history was taken and therapeutic history was taking including need for oral corticosteroid, frequency of use and the response to it.2- Complete clinical examination including general, respiratory tract and systemic examinations.3- Pulmonary function tests were performed in chest department by pulmonologist. Spirometry was carried out on a HypAir compact plus flowmeter pulmonary function testing station (Medisoft, Solinnes, Belgium) in sitting position with nose clip following recommendations of European respiratory society and American respiratory society for test of pulmonary function.4- The forced vital capacity (FVC), the forced expiratory volume in 1st second (FEV1) and the ratio (FEV1/FVC) were measured.5- Spirometric indices were calculated using the best of three technically satisfactory performance [11]. Predicted values of FEV1, FVC based on age, sex, and height [12]. So, it is important that results are compared with normal standards for age, sex, race and height [13].
2.1. Sample Collection
2.1.1. Serum and Blood Samples Collection
- Venous blood samples were collected aseptically from all patients in three separate test tubesa) One sterile serum separator tube was used for measurement of IgE by putting into plain tube without anticoagulants. After coagulation, sample was centrifuged (for 15 minutes at 1000 ×g) and serum was harvested, divided into aliquots and stored at -20°C until analysis. b) Tow sterile tubes containing EDTA were used for complete blood picture, and DNA extraction.
2.1.2. Laboratory Investigations
- I. Complete blood count (CBC) using SYSMEX KX 21-N Cell Counter.II. Serum IgE was detected by ELISA immunoassay Kits from DRG diagnostics, EIA-1788, USA. The DRG IgE Quantitative Test is a solid phase enzymelinked immunosorbent assay (ELISA) based on the sandwich principle. The test specimen (serum) is added to the IgE monoclonal antibodies immobilized on polystyrene microtiter wells (solid phase) and incubated with the Zero Buffer. If human IgE is present in the specimen, it will combine with the antibodies on the well. The well is then washed to remove any residual test specimen, and goat anti-IgE in the antibody-enzyme (horseradish peroxidase) conjugate reagent is added. The conjugate reagent will bind immunologically to the IgE on the well, resulting in the IgE molecules being sandwiched between the solid phase and the enzyme-linked antibodies. After incubation at room temperature, the solid phase is washed with water to remove unbound labeled antibody. A solution of 3,3’,5,5’-Tetramethylbenzidine (TMB) is added and incubated for 20 minutes, resulting in the development of a blue color. The color development is stopped and the resulting yellow color is measured spectrophotometrically at 450 nm. The concentration of IgE is directly proportional to the color intensity of the test sample.III. Genotyping of IL-10 gene polymorphism rs1800896 (-1082 G/A)The IL-10 gene SNP rs 1800896 was selected. SNP genotyping was conducted using the real time polymerase chain reaction (PCR). Real time PCR amplification was done by Rotor-Gene Q Real Time PCR instruments from Qiagen USA, Using real-time PCR kit (Lot Number 6294826, Applied Bio-systems). Total genomic DNA was extracted from peripheral venous blood of each individual using DNA extraction and purification kit (QIAamp DNA Blood Mini Kit, Qiagen, Germany) fully automated on the QIAcube (Germany). QIAamp DNA Blood Mini Kits are designed for rapid purification of an average of 6 μg of total DNA from 200 μl of whole human blood. Applied Biosystems recommends quantifying the amount of genomic DNA in samples before using TaqMan SNP Genotyping Assays. The amount of DNA was measured spectrophotometrically for concentration and purity. All samples had an optical density (OD) 260/280 nm ratio >1.8, indicating high purity. The DNA integrity was tested on the Nanodrop ND-1000 spectrophotometer (Germany) which is a full spectrum (220 -750nm) spectrophotometer that measures 1 μl samples with high accuracy and reproducibility. The SNP Genotyping Assay contains sequence-specific forward and reverse primers to amplify the polymorphic sequence of interest. The following primer pairs were used: forward 5'-CAACACTACTAAGGCTTCTTTGGGA-3' and reverse 5'-GGGGAAGTAGGGATAGGTAAGAGGA-3'. Target DNA sequences were amplified in a total volume of 20 μL, containing 10 μL TaqMan Universal PCR Master Mix, 1.0 μL TaqMan working stock of SNP Genotyping 20X, 9.0 μL DNA sample and Nuclease-free water (Master Mix preparation). Thermal cycling conditions were 10 min at 95°C followed by 50 cycles of 15 sec at 92°C (denature) and 90 sec at 60°C (anneal/extend).
2.2. Statistical Analysis
- Data were analyzed using SPSS version 17.0 (Statistical Package for Social Sciences Inc., Chicago, IL, USA) and Microsoft Excel 2010. Parametric data was expressed as mean ± SD and non-parametric data was expressed as number and percentage. Student’s t test was done to compare between two groups. Chi-square test was used to examine the relationship between two qualitative variables. p value of >0.05 considered non-significant, p value of ≤0.05 considered significant, p value of <0.01 was considered highly significant.
3. Results
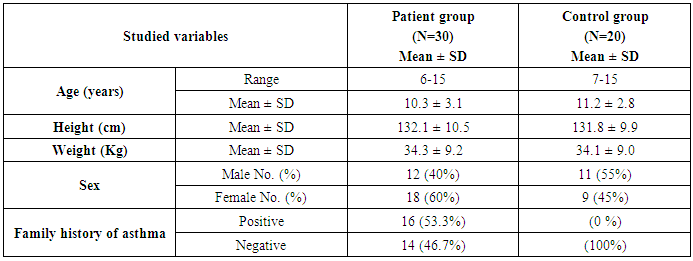
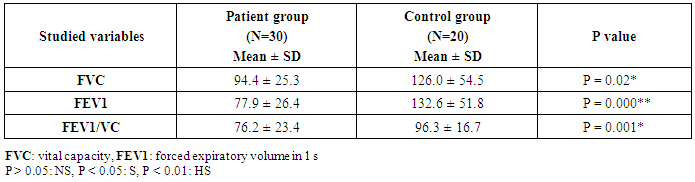
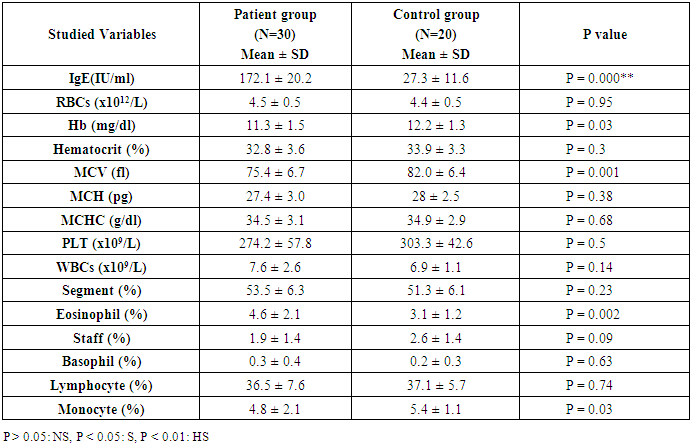
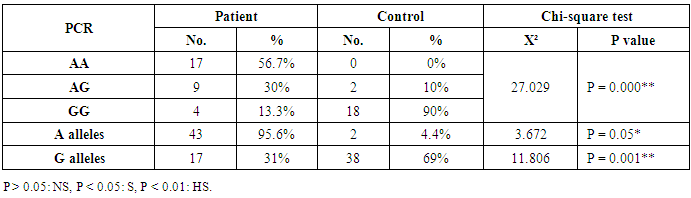
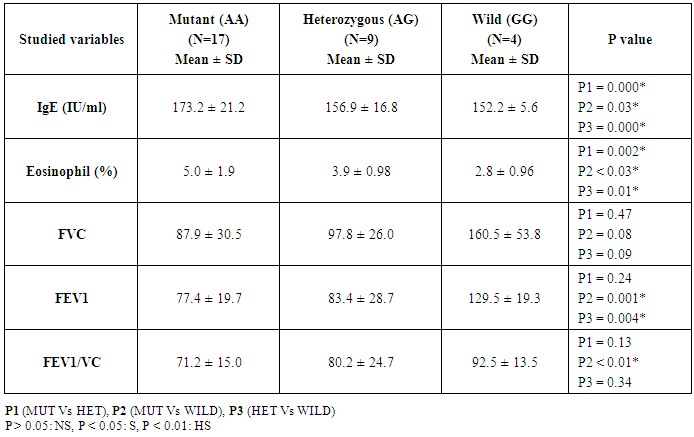
- This study was conducted on thirty children suffering from bronchial asthma, including 12 males and 18 females with an age range of 6 and 15 years old (10.3 ± 3.1). Twenty age and gender matched apparently healthy children was used as control, demographic data of studied groups was presented in table (1).As regard to Pulmonary function tests, there was significantly decreased in vital capacity (VC), forced expiratory volume in 1s (FEV1) and FEV1/VC ratio in asthmatic children when compared to control group (p= 0.000) (Table 2).Laboratory parameters for patient and control group were presented in table (3). As expected, there was highly significant increase in IgE level and Eousenophil (%) in asthmatic children compared to control group (p= 0.000 and 0.002). There was significant decrease in Hb, MCV and monocytes in asthmatic children compared to control group (p= 0.03, 0.002 and 0.03). No significant differences were observed between studied groups regarding other parameters.The Frequency and number of alleles of SNP rs1800896 (-1082 G/A) in asthmatic children compared to control group were presented in table (4). The frequency of SNP rs 1800896 showed that the homozygous mutant type (AA) was detected in (56.7%) of asthmatic children and (0%) of control & the heterozygous mutant type (AG) was detected in (30%) of asthmatic children and (10%) of control while wild type (GG) was detected in (13.3%) of asthmatic children and (90%) of control with statistically high significant differences between both groups. The number of A allele was increased in asthmatic children (43) as compared with (2) A alleles in control group, while G allele was increased in control group (38) as compared with (17) G alleles in asthmatic children.Clinical and Lab characteristics according to the IL-10 promoter polymorphisms within asthmatics were presented in table (5). There was significant increase in serum IgE and eosinophil count in homozygous mutan (AA) and heterozygous (AG) than wild type (GG) (p= 0.03, p= 0.000). There was significant decrease in respiratory functions FEV1, FVC/FEV1 in homozygous mutant (AA) and heterozygous (AG) than wild type (GG) (p= 0.001, p= 0.01).
|
|
|
|
|
4. Discussion
- Pediatric asthma is one of the most common diseases affecting children worldwide. According to the World Health Organization (WHO), approximately 235 million people suffered from asthma worldwide, with 157,000 requiring urgent medical aid, during 2007 [14].The onset of asthma occurs before the age of 12 (years) in over half the cases. The past 20 years have seen a significant increase in the prevalence of asthma in children. The pathogenesis of asthma is related to multiple risk factors including environmental factors such as vehicular pollution, and genetic factors such as polymorphisms in the interleukin IL-4 and ORMDL3 genes [15-17].Pulmonary function tests are non-invasive tests, used to detect air flow limitation and/or lung volume restriction. Assessment of ventilator function is an important investigation because early detection of functional impairment and its appropriate treatment will help to reduce morbidity and mortality related to disease [18].Present study reported that there was significantly decreased in forced vital capacity (FVC), forced expiratory volume in 1s (FEV1) and FEV1/VC ratio in asthmatic children when compared to control group (p= 0.000), which in consistent with previous studies that showed decreased lung function parameters in asthma patients as compared to control group [18, 19].Current study demonstrated that there was highly significant increase in IgE level in asthmatic children compared to control group (p= 0.000). These results were similar to Smolnikova et al. [1], they found over production of IgE in both controlled bronchial asthma and uncontrolled bronchial asthma as compared with the control.Regarding to Eosenophil (%), present study found significant increase in Eosenophil (%) in asthmatic children compared to control group (p= 0.002), which in agreement of Volbeda et al. [20], they shown that patients with uncontrolled asthma exhibit higher eosinophil numbers in peripheral blood.A number of studies have reported that IL-10 plays an important role in the pathogenesis of asthma [21]. IL-10 is an important anti-inflammatory factor secreted by Th2 cells, monocytes/macrophages, and B lymphocytes. IL-10 stimulates the proliferation and activation of B lymphocytes, in addition to inhibiting the synthesis of TNF-γ and IL-2. Studies have also suggested that IL-10 could reduce allergic inflammation by inhibiting the production of pro-inflammatory cytokines [22].IL-10 is located at chromosome 1q31-q32, a region associated with asthma susceptibility. Previous studies assessing the association between IL-10 promoter polymorphisms and susceptibility to asthma have yielded inconsistent and inconclusive results. A majority of these studies have focused on single nucleotide polymorphisms in the promoter region -1082G/A (rs1800896) [23-25].One of the major goals of SNP studies is to perceive the genetic basis of human phenotype variation, especially of complex diseases. Identification of asthma-associated genetic variants may be clinically useful for identification of the patients at risk [8].Our results showed an association between IL-10 rs1800896 SNPs and bronchial asthma in Egyptian population as we found that the homozygous mutant type (AA) -1082 G/A (rs1800896) was detected in (56.7%) of asthmatic children and (0%) of control & the heterozygous mutant type (AG) was detected in (30%) of asthmatic children and (10%) of control, while wild type (GG) was detected in (13.3%) of asthmatic children and (90%) of control with the statistically highly significant differences between both groups (p= 0.000).These results were consistent with previous studies which indicated that -1082 G/A (rs1800896) polymorphism might be associated with increased susceptibility to asthma [26, 27]. Hussein et al. [28] also reported that the -1082 AA genotype increased asthma risk in Egyptian children. As well as, Xu et al. [29] reported a similar finding in a Chinese population.Both Nie et al. [30] and Zheng et al. [31] were supported our finding as they concluded that the IL-10 -1082G/A polymorphism was significantly associated with asthma susceptibility.In contrast, other studies have reported no significant association between -1082 G/A (rs1800896) polymorphism and the risk of pediatric asthma; Kim et al. [32] revealed that the IL-10 -1082G/A polymorphism were not associated with pediatric asthma in a Korean population, which was consistent with the results obtained by Li et al. [33].Current study revealed that number of A allele at position -1082 of the Il-10 gene was increased in asthmatic children (43) as compared with (2) A alleles in control group, while G allele was increased in control group (38) as compared with (17) G alleles in asthmatic children.These results were in agreement of Chatterjee et al. [34], they found that The A allele at position -1082 of the Il-10 gene, which determines an impaired secretion of IL-10 in asthma patients, was more frequent in patients with asthma than in the control group in a study conducted on Indians.Similar to our findings, Smolnikova et al. [1] reported that G allele in the -1082 position correlates with high production of IL-10 as compared to the alternative allele A-1082.In contrast, Zedan et al. [27] reported that the GG genotype at the IL-10 -1082 region could be a contributing factor for the increased susceptibility to, and severity of, childhood asthma in Egypt.Trajkov et al. [35] analyzed the role of 22 cytokine polymorphisms in 74 patients with asthma and 301 controls and reported no correlation between a specific allele at position -1082 and asthma. However, genotype analysis revealed a positive association between GG and AA genotypes and asthma in Macedonians [34]. Karjalainen et al. [36] reported no correlation between this alleles and asthma susceptibility in Finnish.The present study demonstrated that there was significant increase in serum IgE and eosinophil count in homozygous mutant (AA) and heterozygous (AG) than wild type (GG) (p= 0.03, p= 0.000), indicating that A allele associated with excess production of IgE in asthmatic children, which in agreement of Smolnikova et al. [1], they shown that the G -1082 A polymorphism of IL10 along with the ATA haplotype in the promoter of IL10 are associated with enhanced bronchial hyperresponsiveness.In contrast, Kim et al. [32] observed no correlation between IL-10 -1082 genotypes and IL-10 levels, total IgE levels, eosinophil count or asthma susceptibility.Regarding to respiratory functions, present study showed that there was significant decrease in respiratory functions FEV1, FVC/FEV1 in homozygous mutant (AA) and heterozygous (AG) than wild type (GG) (p= 0.001, p= 0.01), which in consistence with previous studies reported that, there was a correlation between the A allele and FEV1 reduction, indicating an association with severe types of asthma in children [37, 38].Similarly, there was also a correlation between the AA genotype and the reduced pulmonary function observed in Kim’s study [32].In contrast, Karjalainen et al. [36] found no association between IL-10 polymorphism and lung function in asthmatic patients. As lung function is a complex trait, a couple of genes may not cause enough variation to change the spirometric parameters significantly.
5. Conclusions
- The results of this study found that the IL-10 -1082 G/A polymorphism was associated with increased susceptibility to pediatric asthma and related to reduced respiratory functions between asthmatics. Further large scale investigations are required to assess the potential role of this polymorphism in bronchial asthma.
Limitations of the Current Study
- Included the relatively small number of enrolled patient and the genotyping of only one SNP (SNP rs1800896). So, further studies on larger number of patients are required to confirm the current findings & to detect the genetic susceptibility to Bronchial Asthma in Egyptians.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML